Volume 15, Number 12—December 2009
Dispatch
Sympatry of 2 Hantavirus Strains, Paraguay, 2003–2007
Abstract
To explore geographic and host-taxonomic patterns of hantaviruses in Paraguay, we established sampling sites in the Mbaracayú Biosphere Reserve. We detected Jaborá virus and Itapúa37/Juquitiba–related virus in locations ≈20 m apart in different years, which suggested sympatry of 2 distinct hantaviruses.
Hantaviruses are rodent-borne viruses that may cause hemorrhagic fever with renal syndrome or hantavirus pulmonary syndrome in humans, although some strains do not cause disease (1,2). In Paraguay in 1995, Laguna Negra virus carried by Calomys laucha (little laucha) caused an outbreak of hantavirus pulmonary syndrome in western Paraguay (3).
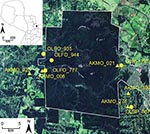
We have identified 4 additional strains in Paraguay: Alto Paraguay virus harbored by Holochilus chacarius (Chacoan marsh rat) in western Paraguay; and Ape Aime virus (AAIV) harbored by Akodon montensis (Montane akodont), Itapúa virus strain 37 (IPV37) harbored by Oligoryzomys nigripes (black-footed colilargo), and Bermejo virus strain Ñeembucu harbored by O. chacoensis (Chacoan colilargo) in eastern Paraguay (4,5). We have continued our surveillance of hantaviruses in the interior Atlantic forests within and near Reserva Natural del Bosque Mbaracayú (RNBM), a World Biosphere Reserve located within Departamento Canindeyú in eastern Paraguay (Figure 1).
We established and sampled 10 mark-recapture sites within and adjacent to RNBM during 2003–2007. Sampling of grids depended upon weather conditions, the purpose of the grid, and transitory human settlements. Each mark-recapture grid consisted of an 11 × 11 array of trap stations spaced 10 m apart, each of which had 1 standard live trap (H.B. Sherman Traps, Tallahassee, FL, USA) placed on the ground, and another in branches or vines 2–3 m above ground to capture arboreal species. Grids were sampled for 8 nights, with at least 2 months between sampling sessions. Rodents captured in the mark-recapture grids were individually marked with passive integrated transponder tags, and ≈100 μL of blood was collected from the retroorbital sinus (once per trapping session). Animals were identified to species, age class, reproductive condition, sex, and weight and released.
Rodents were also collected in a series of traplines, each of which contained 50 traps placed ≈10 m apart. Animals collected in traplines were killed, standard collecting information was recorded, and liver, lung, heart, kidney, muscle tissues, and blood specimens were collected. All samples were snap-frozen in liquid nitrogen, transported to the Museum of Texas Tech University (TTU), and stored at –80°C. Standard voucher specimens were prepared from these animals, and have or will be deposited in the Museum of TTU or the Museo Nacional de Historia Natural del Paraguay. All field protocols followed American Society of Mammologists guidelines for the use of wild mammals in research (6), and were reviewed and approved by the TTU Animal Care and Use Committee.
A total of 1,150 small mammals from >20 species were captured, including 13 sigmodontine rodent species (1,140 animals) (Technical Appendix). The dominant rodent species in Mbaracayú were A. montensis (55.7%), Necromys lasiurus (hairy-tailed akodont) (10.8%), C. callosus (big laucha) (6.5%), and O. fornesi (Fornes’ colilargo) (6.3%).
Antibodies to hantavirus antigens were detected in blood specimens by using an indirect immunofluorescent antibody assay and irradiation-sterilized slides of Vero E6 cells infected with Andes virus as described (4). Seven species were antibody positive: A. montensis, N. lasiurus, O. fornesi, O. nigripes, Oligoryzomys sp., Oryzomys megacephalus (Azara’s broad-headed Oryzomys sp.), and Oxymycterus delator (Paraguayan hocicudo). Antibodies to hantavirus antigens were 3× more abundant in blood samples from males than females (Technical Appendix).
Total RNA was extracted from antibody-positive blood clots from mark-recapture samples or lung tissue from killed animals. Nested reverse transcription–PCR was performed to amplify a 371-nt small (S) hantavirus RNA segment (4). Hantavirus RNA was detected in 23 A. montensis, 5 O. fornesi, 1 O. nigripes, and 1 Oligoryzomys sp. Of these animals, all but 2 A. montensis were males, which indicated that male rodents play the primary role in maintenance and transmission of hantavirus.
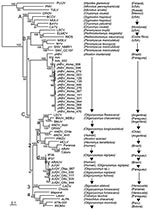
A representative sample (15 A. montensis, 3 O. fornesi, 1 O. nigripes, and 1 Oligoryzomys sp.) were selected for additional PCR, sequencing, and phylogenetic analysis. PCR-amplified cDNAs of a 1,014-nt amino terminus region of the S segment were purified by agarose gel electrophoresis and cloned into pCRII (Invitrogen, Carlsbad, CA, USA) (4,5). M13 forward and reverse primers were used for sequencing. For sequence comparison and phylogenetic analysis, sequences of representative New and Old World hantaviruses were obtained from GenBank. Phylogeny reconstruction was conducted by using Modeltest version 3.6 analysis (http://darwin.uvigo.es/software/modeltest.html.), maximum-likelihood estimation, and Bayesian inference (Figure 2).
Bayesian analysis based on the 1,014-nt sequence showed that all sequences from A. montensis formed a strongly supported clade, which included AAIV-related hantaviruses from Itapúa Department, Jaborá virus (JABV) from southern Brazil, and strains from RNBM in Paraguay (Figure 2, clade C1). Phylogenetic analyses identified 3 subclades representing virus sequences from animals in the RNBM, Itapúa, and southern Brazil. This type of geographic clustering is similar to Sin Nombre–related viruses in deer mice in North America (7). All S segment sequences from A. montensis were closely related, with nucleotide sequence differences between RNBM strains and AAIV and JABV of 4% and 12%, respectively, and derived amino acid differences of 0% or 1%, respectively (Table 1).
In contrast, all virus sequences from O. fornesi, O. nigripes, and Oryzomys sp. at RNBM formed a strongly supported clade with viruses related to Juquitiba virus (JUQV) from Brazil and Itapúa virus strain 37, which was originally detected in O. nigripes from Itapúa Department in eastern Paraguay (Figure 2, clade C2d). Nucleotide sequence differences between JUQV strains from RNBM were 0%–2%. Nucleotide sequence differences between JUQV strains from RNBM and Itapúa virus strain 37 or JUQV (Brazil) were 5% or 4%, respectively, and derived amino acid differences were 0% (Table 1). This clade is phylogenetically distinct from viruses that form the Akodon montensis clade at RNBM and more closely related to Andes (clade C2b) and Bermejo-Ñeembucú (clade C2a) viruses. This finding suggests that spillover infection of JUQV-related viruses is actively occurring among oryzomyine rodent species at RNBM, as reported for other hantaviruses in oryzomyines (8) and other rodent hosts of Old World hantaviruses (9,10). Additional data are needed to determine the primary oryzomyine reservoir of JUQV and to better understand mechanisms by which spillover occurs.
In addition to spillover infection of JUQV among oryzomyine rodents, we identified 2 virus strains (JUQV and JABV) in close proximity (collected ≈20 m apart on the same grid in the same sampling session) on 2 occasions, in sites separated by ≈30 km (Figure 1; Table 2). Thus, these 2 distinct hantaviruses appear to be maintaining a sympatric status across a considerable expanse of landscape, rather than reflecting a temporary or localized phenomenon. We use the term sympatric to underscore that these viruses are in the same community and are near enough (their rodent reservoirs) to interact.
Coexistence of hantaviruses in 2 rodent species at mark-recapture sites has been observed (11–13). Serologic analyses in these studies would not have differentiated whether distinct strains of hantaviruses were co-circulating or active spillover infection was occurring among sympatric rodents at collection sites. Recently, Raboni et al. reported JUQV circulating in 3 sympatric rodent species in southern Brazil and 2 distinct hantaviruses (Jabora and JUQV) in 1 rodent species (A. montensis) (14). We have not detected JUQV in A. montensis in Paraguay. To address host-jumping of hantaviruses among sympatric rodent species in RNBM and other regions in South America, future longitudinal studies are warranted. Such studies are critical to understanding evolutionary adaptation of hantaviruses in rodents in South America, the ability of these viruses to adapt to new rodent reservoirs, and their emergence and maintenance in the environment.
Dr Chu is a research scientist at Southern Research Institute. His research interests include the ecology and evolution of hantaviruses.
Acknowledgments
We thank Robert J. Baker and Heath Garner for approving and facilitating loans of rodent tissues; the Secretaría del Ambiente (Paraguay) for providing permits to collect and export rodents and tissues; and the field crew, led by Ismael Mora, for dedication to their work, regardless of circumstances.
This work was supported by a grant from the Fogarty International Center (1 R01 TW006986-01) to C.B.J. under the National Institutes of Health–National Science Foundation Ecology of Infectious Diseases initiative.
References
- Schmaljohn C, Hjelle B. Hantaviruses: a global disease problem. Emerg Infect Dis. 1997;3:95–104. DOIPubMedGoogle Scholar
- Vapalahti O, Mustonen J, Lundkvist A, Henttonen H, Plyusnin A, Vaheri A. Hantavirus infections in Europe. Lancet Infect Dis. 2003;3:653–61. DOIPubMedGoogle Scholar
- Johnson AM, Bowen MD, Ksiazek TG, Williams RJ, Bryan RT, Mills JN, Laguna Negra virus associated with HPS in western Paraguay and Bolivia. Virology. 1997;238:115–27. DOIPubMedGoogle Scholar
- Chu YK, Owen RD, Gonzalez LM, Jonsson CB. The complex ecology of hantavirus in Paraguay. Am J Trop Med Hyg. 2003;69:263–8.PubMedGoogle Scholar
- Chu YK, Milligan B, Owen RD, Goodin DG, Jonsson CB. Phylogenetic and geographical relationships of hantavirus strains in eastern and western Paraguay. Am J Trop Med Hyg. 2006;75:1127–34.PubMedGoogle Scholar
- Gannon WL, Sikes RS; Animal Care and Use Committee of the American Society of Mammalogists. Guidelines of the American Society of Mammalogists for the use of wild mammals in research. J Mammal. 2007;88:809–23. DOIGoogle Scholar
- Monroe MC, Morzunov SP, Johnson AM, Bowen MD, Artsob H, Yates T, Genetic diversity and distribution of Peromyscus-borne hantaviruses in North America. Emerg Infect Dis. 1999;5:75–86. DOIPubMedGoogle Scholar
- Torrez-Martinez N, Bharadwaj M, Goade D, Delury J, Moran P, Hicks B, Bayou virus-associated hantavirus pulmonary syndrome in eastern Texas: identification of the rice rat, Oryzomys palustris, as reservoir host. Emerg Infect Dis. 1998;4:105–11. DOIPubMedGoogle Scholar
- Klingstrom J, Heyman P, Escutenaire S, Sjölander KB, De Jaegere F, Henttonen H, Rodent host specificity of European hantaviruses: evidence of Puumala virus interspecific spillover. J Med Virol. 2002;68:581–8. DOIPubMedGoogle Scholar
- Weidmann M, Schmidt P, Vackova M, Krivanec K, Munclinger P, Hufert FT. Identification of genetic evidence for Dobrava virus spillover in rodents by nested reverse transcription (RT)-PCR and TaqMan RT-PCR. J Clin Microbiol. 2005;43:808–12. DOIPubMedGoogle Scholar
- Calisher CH, Root JJ, Mills JN, Rowe JE, Reeder SA, Jentes ES, Epizootiology of Sin Nombre and El Moro Canyon hantaviruses, southeastern Colorado, 1995–2000. J Wildl Dis. 2005;41:1–11.PubMedGoogle Scholar
- Artois M, Cochez C, Mele RV, Heyman P. Genetic evidence of Puumala and Tula hantaviruses in rodents in the Jura region, France: preliminary results. Eurosurveillance. 2007;12:E070628.3.
- Mills JN, Schmidt K, Ellis BA, Calderon G, Enria DA, Ksiazek TH. A longitudinal study of hantavirus infection in three sympatric reservoir species in agroecosystems on the Argentine pampa. Vector Borne Zoonotic Dis. 2007;7:229–40. DOIPubMedGoogle Scholar
- Raboni SM, Hoffmann FG, Oliveira RC, Teixeira BR, Bonvicino CR, Stella VS, Phylogenetic characterization of hantaviruses from wild rodents and hantavirus pulmonary syndrome cases in the state of Parana (southern Brazil). J Gen Virol. 2009;90:2166–71. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 15, Number 12—December 2009
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Colleen B. Jonsson, Department of Microbiology and Immunology, Center for Predictive Medicine for Biodefense and Emerging Infectious Diseases, University of Louisville, Louisville, KY 40222, USA
Top