Volume 15, Number 12—December 2009
Dispatch
Molecular Epidemiology of Glanders, Pakistan
Figure 1
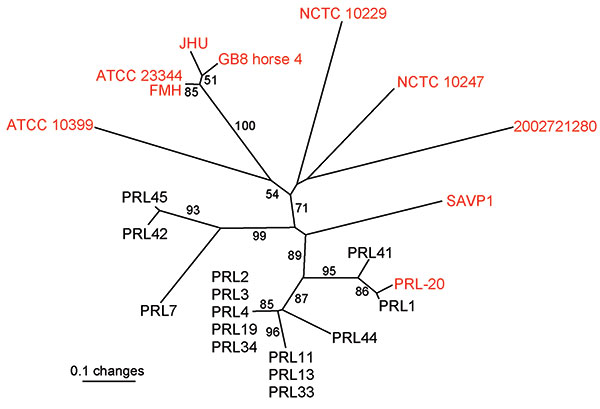
Figure 1. Unrooted neighbor-joining tree based on 23 variable number tandem repeat loci demonstrating that the Punjab isolates (black text and PRL-20) are genetically distinct from and less diverse than available whole genome sequences (WGS, red text) (8). Statistical supports for branches based on 1,000 bootstrap iterations are shown. Sample PRL-20 is shown in red text because it is also available as a whole genome sequence; therefore, it was used in all 3 situations where an average pairwise distance (APD) was calculated. Among 10 WGS, the average pairwise distance was 0.607; between 10 WGS and Punjab isolates, average pairwise distance was 0.627; and among 15 Punjab isolates, average pairwise distance was 0.312. These results indicate that the Punjab isolates are more closely related to each other than to the sequenced strains because the APD among Punjab isolates is 2× lower than the APD calculated in the other 2 situations.
References
- M’Fadyean J. Glanders. J Comp Pathol Ther. 1904;17:295–317.
- Waag DM, DeShazer D. Glanders: new insights into an old disease. In: Lindler LE, Lebeda FJ, Korch GW, editors. Biological weapons defense: infectious diseases and counterbioterrorism. 1st ed. Totowa (NJ): Humana Press Inc.; 2004. p. 209–37.
- Gilad J, Harary I, Dushnitsky T, Schwartz D, Amsalem Y. Burkholderia mallei and Burkholderia pseudomallei as bioterrorism agents: national aspects of emergency preparedness. Isr Med Assoc J. 2007;9:499–503.PubMedGoogle Scholar
- Naureen A, Saqib M, Muhammad G, Hussain MH, Asi MN. Comparative evaluation of Rose Bengal plate agglutination test, mallein test, and some conventional serological tests for diagnosis of equine glanders. J Vet Diagn Invest. 2007;19:362–7.PubMedGoogle Scholar
- Michelle Wong Su Y, Lisanti O, Thibault F, Toh Su S, Loh Gek K, Hilaire V, . Validation of ten new polymorphic tandem repeat loci and application to the MLVA typing of Burkholderia pseudomallei isolates collected in Singapore from 1988 to 2004. J Microbiol Methods. 2009 Mar 25.
- Moore DD. Preparation and analysis of DNA. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, et al., editors. Current protocols in molecular biology. New York: Wiley & Sons; 1995. p. 2.4.1–2.4.2.
- U’Ren JM, Schupp JM, Pearson T, Hornstra H, Friedman CL, Smith KL, et al. Tandem repeat regions within the Burkholderia pseudomallei genome and their application for high resolution genotyping. BMC Microbiol. 2007;7:23. DOIPubMedGoogle Scholar
- NCBI. ENTREZ Genome Project. Burkholderia mallei [cited 2009 Jul 21]. Available from http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=search&term=txid13373
- Pearson T, U’Ren JM, Schupp JM, Allan GJ, Foster PG, Mayo MJ, et al. VNTR analysis of selected outbreaks of Burkholderia pseudomallei in Australia. Infect Genet Evol. 2007;7:416–23. DOIPubMedGoogle Scholar
- Girard JM, Wagner DM, Vogler AJ, Keys C, Allender CJ, Drickamer LC, Differential plague-transmission dynamics determine Yersinia pestis population genetic structure on local, regional, and global scales. Proc Natl Acad Sci U S A. 2004;101:8408–13. DOIPubMedGoogle Scholar
- Godoy D, Randle G, Simpson AJ, Aanensen DM, Pitt TL, Kinoshita R, Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol. 2003;41:2068–79. DOIPubMedGoogle Scholar
- Pakistan Agriculture Census Organization, Government of Pakistan, Statistics Division. Pakistan livestock census. 2006 [cited 2009 Apr 15]. Available from http://www.statpak.gov.pk/depts/aco/publications/pakistan-livestock-cencus2006/lsc2006.html
- Al-Ani FK, Al-Delaimi AK, Ali AH. Glanders in horses: clinical and epidemiological studies in Iraq. Pakistan Vet J. 1987;7:126–9.
- Henning MW. Glanders, farcy, droes, malleus. Animal diseases in South Africa being an account of the infectious diseases of domestic animals. Third edition (completely revised) including a description of a number of diseases not reported in the previous edition. Johannesburg (South Africa): Central News Agency Ltd.; 1956.
- Miller WR, Pannell L, Cravitz L, Tanner WA, Ingalls MS. Studies on certain biological characteristics of Malleomyces mallei and Malleomyces pseudomallei: I. morphology, cultivation, viability, and isolation from contaminated specimens. J Bacteriol. 1948;55:115–26.