Volume 15, Number 3—March 2009
Dispatch
Detection of Newly Described Astrovirus MLB1 in Stool Samples from Children
Figure 2
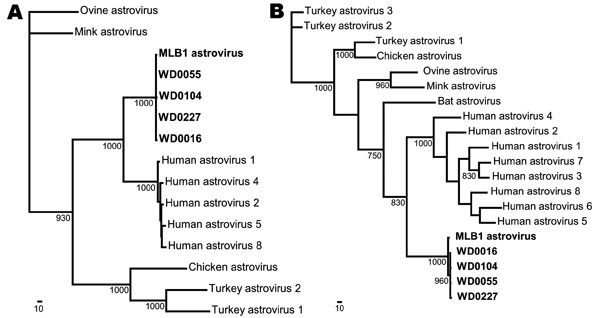
Figure 2. Phylogenetic analysis of astrovirus MLB1 (AstV-MLB1) isolates. A region of the serine protease (A) and the capsid (B) of each virus detected by the AstV-MLB1–specific primers was amplified and sequenced. Multiple sequence alignments were then generated with these sequences and the corresponding regions of known astroviruses using ClustalX (www.clustal.org). PAUP* (Sinauer Associates, Sunderland, MA, USA) was used to generate phylogenetic trees; bootstrap values (>700) from 1,000 replicates are shown. The previously identified AstV-MLB1 isolate (9,10) and the isolates from this study are shown in boldface. Scale bars indicate number of amino acid substitutions per site.
References
- Chu DK, Poon LL, Guan Y, Peiris JS. Novel astroviruses in insectivorous bats. J Virol. 2008;82:9107–14. DOIPubMedGoogle Scholar
- Koci MD, Schultz-Cherry S. Avian astroviruses. Avian Pathol. 2002;31:213–27. DOIPubMedGoogle Scholar
- Moser LA, Schultz-Cherry S. Pathogenesis of astrovirus infection. Viral Immunol. 2005;18:4–10. DOIPubMedGoogle Scholar
- Caracciolo S, Minini C, Colombrita D, Foresti I, Avolio M, Tosti G, Detection of sporadic cases of Norovirus infection in hospitalized children in Italy. New Microbiol. 2007;30:49–52.PubMedGoogle Scholar
- Glass RI, Noel J, Mitchell D, Herrmann JE, Blacklow NR, Pickering LK, The changing epidemiology of astrovirus-associated gastroenteritis: a review. Arch Virol Suppl. 1996;12:287–300.PubMedGoogle Scholar
- Kirkwood CD, Clark R, Bogdanovic-Sakran N, Bishop RF. A 5-year study of the prevalence and genetic diversity of human caliciviruses associated with sporadic cases of acute gastroenteritis in young children admitted to hospital in Melbourne, Australia (1998–2002). J Med Virol. 2005;77:96–101. DOIPubMedGoogle Scholar
- Klein EJ, Boster DR, Stapp JR, Wells JG, Qin X, Clausen CR, Diarrhea etiology in a children’s hospital emergency department: a prospective cohort study. Clin Infect Dis. 2006;43:807–13. DOIPubMedGoogle Scholar
- Soares CC, Maciel de Albuquerque MC, Maranhao AG, Rocha LN, Ramirez ML, Benati FJ, Astrovirus detection in sporadic cases of diarrhea among hospitalized and non-hospitalized children in Rio de Janeiro, Brazil, from 1998 to 2004. J Med Virol. 2008;80:113–7. DOIPubMedGoogle Scholar
- Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog. 2008;4:e1000011. DOIPubMedGoogle Scholar
- Finkbeiner SR, Kirkwood CD, Wang D. Complete genome sequence of a highly divergent astrovirus isolated from a child with acute diarrhea. Virol J. 2008;5:117.PubMedGoogle Scholar
- Noel JS, Lee TW, Kurtz JB, Glass RI, Monroe SS. Typing of human astroviruses from clinical isolates by enzyme immunoassay and nucleotide sequencing. J Clin Microbiol. 1995;33:797–801.PubMedGoogle Scholar
- Swofford DL. PAUP*: phylogenetic analysis using parsimony (*and other methods). Version 4. Sunderland (MA): Sinauer Associates; 1998.
- Chiu CY, Greninger AL, Kanada K, Kwok T, Fischer KF, Runckel C, Identification of cardioviruses related to Theiler's murine encephalomyelitis virus in human infections. Proc Natl Acad Sci U S A. 2008;105:14124–9. DOIPubMedGoogle Scholar
Page created: December 07, 2010
Page updated: December 07, 2010
Page reviewed: December 07, 2010
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.