Volume 15, Number 4—April 2009
Research
Exotic Small Mammals as Potential Reservoirs of Zoonotic Bartonella spp.
Abstract
To evaluate the risk for emerging human infections caused by zoonotic Bartonella spp. from exotic small mammals, we investigated the prevalence of Bartonella spp. in 546 small mammals (28 species) that had been imported into Japan as pets from Asia, North America, Europe, and the Middle and Near East. We obtained 407 Bartonella isolates and characterized them by molecular phylogenetic analysis of the citrate synthase gene, gltA. The animals examined carried 4 zoonotic Bartonella spp. that cause human endocarditis and neuroretinitis and 6 novel Bartonella spp. at a high prevalence (26.0%, 142/546). We conclude that exotic small mammals potentially serve as reservoirs of several zoonotic Bartonella spp.
The genus Bartonella includes a variety of gram-negative, fastidious, hemotrophic bacteria that are transmitted by blood-sucking arthropod vectors. The genus consists of 20 species and 3 subspecies; at least 11 of these species are known or suspected to be pathogenic for humans as causative agents of emerging zoonoses (1).
The following Bartonella spp. have been isolated from wild mice: B. birtlesii (2), B. doshiae, B. grahamii, B. taylorii (3), and B. vinsonii subsp. arupensis and subsp. vinsonii (4). In several countries, the following species have been carried by rats of the genus Rattus: B. elizabethae (5), B. tribocorum (6), B. phoceensis, and B. rattimassiliensis (7). In South Africa, strains genetically related to B. elizabethae also have been isolated from mice of the genera Aethomys and Tatera (8). The main reservoir of B. washoensis is considered to be wild squirrels (9). Of these rodent-associated Bartonella spp., B. elizabethae, B. grahamii, B. vinsonii subsp. arupensis, and B. washoensis have been implicated in the human infections endocarditis (10), neuroretinitis (11), pyrexia and endocarditis (4,12), and myocarditis (9), respectively.
Previous studies have demonstrated high prevalence of infection with Bartonella spp. in wild and peridomestic small animals in Europe (7,13–15), North and South America (5,16–19), Asia (20–23), and Africa (8). Thus, these animals are thought to be reservoirs of several Bartonella spp. and sources of infection for humans.
Many exotic animals are traded as pets around the world and have been imported into Japan without quarantine. However, no data exist on the prevalence of infection with Bartonella spp., especially in exotic pet animals. Our study objectives were to 1) examine the prevalence of Bartonella spp. infection in exotic small mammals imported into Japan from various countries, 2) compare the diversity of these Bartonella strains by analyzing the partial sequence of the citrate synthase gene (gltA), and 3) evaluate the possibility that these mammals may serve as potential reservoirs of zoonotic Bartonella spp.
Animals and Samples
For this study, 546 exotic small mammals were purchased from trading companies. The animals represented 3 orders and included 6 families, 23 genera, and 28 species (Table 1). They had been imported into Japan as pets from June 2004 through October 2007 from 8 countries in 4 geographic regions: Asia (China, Thailand, and Indonesia), Europe (the Netherlands and Czech Republic), North America (United States), and the Middle and Near East (Egypt and Pakistan). Of the 546 animals, 367 had been captured in their natural environment and 179 had been bred in the exporting countries. Heparinized blood samples were aseptically collected from each animal (anesthetized with chloroform) and centrifuged at 3,000 rpm for 15 min. Plasma was removed and the blood sample pellets were kept at –80ºC until examination.
Isolation of Bacteria
The blood sample pellets were thawed at room temperature, 100-μL supplemented Medium 199 (24) was added to each pellet, and each sample was mixed well. A 100-μL sample of each mixture was plated on 2 heart infusion agar (DIFCO, Sparks Glencoe, MI, USA) plates containing 5% defibrinated rabbit blood. The plates were incubated at 35ºC under 5% CO2. After 2 weeks of incubation, 2 or 3 colonies with genus Bartonella morphologic characteristics (small, gray or cream-yellow, round colonies) were picked from each plate, confirmed to be gram-negative pleomorphic bacteria, and subcultured using the same conditions used for the original cultures.
DNA Extraction and PCR
The genomic DNA of each isolate was extracted by using InstaGene Matrix (Bio-Rad, Hercules, CA, USA). The extracted DNA was used for PCR analysis of a 312-bp part of the gltA gene to confirm that the bacteria were from the genus Bartonella. PCR was performed by using an iCycler (Bio-Rad) with a 20-μL mixture containing 20 ng extracted DNA, 200 μM of each deoxynucleoside triphosphate, 1.5 mmol/L MgCl2, 0.5 U Taq DNA polymerase (Promega, Madison, WI, USA), and 1 pmol of each primer. The specific primer pair and PCR conditions for gltA were as previously reported (25).
DNA Sequencing and Phylogenetic Analysis
The PCR products were purified by using a commercial kit (Spin Column PCR Products Purification Kit; Bio Basic, Markham, Ontario, Canada). Direct DNA sequencing of the purified PCR products was carried out by using dye terminator chemistry with specific primers (25) and a Model 3130 Genetic Analyzer (Applied Biosystems, Foster City, CA, USA). The 312-bp gltA sequences from the isolates and type strains of established Bartonella spp. in GenBank/EMBL/DDBJ were aligned with the Clustal X program (26), and a phylogenetic tree was drawn, based on the sequence data and using the neighbor-joining method (27) with the Kimura 2-parameter distance method (28) in MEGA 4 (29). Bootstrap analysis was carried out with 1,000 replications (30).
Statistical Analysis
The results were analyzed in 2×2 tables. Chi-square tests were used to examine the statistical significance; p<0.05 was considered significant.
Prevalence of Bartonellae
The prevalence of bartonellae in the exotic small mammals examined was 26.0% (142/546). A total of 407 isolates were obtained from the 142 bacteremic animals (Table 1). The prevalence by animal origin was 37.3% (137/367) in captive animals and 2.8% (5/179) in animals from breeder facilities. A significantly higher prevalence of bartonellae was observed in captive animals than in animals from breeder facilities (p<0.001). In the captive animals, the prevalence by region varied up to 47.2% in Asia, which is higher than the 39.7% prevalence in North America. The prevalence of bartonellae by corresponding taxonomic family of host animal ranged from 38.6% (49/127) in the family Muridae to 43.9% (69/157) in the family Sciuridae. No bartonellae were detected in animals in the families Octodontidae and Erinaceidae. Among animals from breeders, only 5 chipmunks (Tamias sibiricus) from China were found to be infected with bartonellae; no bartonellae were isolated from animals in the families Petauridae, Muridae, Octodontidae, or Dipodidae.
Bartonellae were isolated from 17 of the 28 animal species studied (Table 1). The prevalence by animal species varied from 9.7% (3/31) in the Cairo spiny mouse (Acomys cahirinus) to 100% (10/10) in the bushy-tailed jird (Sekeetamys calurus). Prevelances were considerably higher for the bushy-tailed jird, large Egyptian gerbil (Gerbillus pyramidum), greater Egyptian jerboa (Jaculus orientalis), and lesser Egyptian jerboa (J. jaculus) at 100% (10/10), 90.0% (9/10), 81.3% (13/16), and 75.0% (6/8), respectively.
DNA Sequences and Phylogeny of Isolates
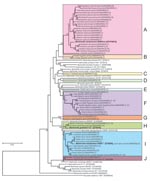
The 407 isolates in this study were classified into 53 genotypes on the basis of DNA sequence analysis of a 312-bp fragment of their gltA genes. The sequence of a genotype from a Cairo spiny mouse was identical to that of the B. elizabethae type strain (GenBank accession no. Z70009) isolated from a human patient with endocarditis (10). The other 52 genotypes were found to be novel genotypes after comparison with known Bartonella spp. The phylogenetic tree of the gltA sequences shows that the 52 novel genotypes are clearly clustered in 10 genogroups, designated A to J (Figure).
Of the 52 novel genotypes, genogroup A, which consisted of 21 genotypes (AB444954 to AB444974) isolated from 7 squirrel species, was related to B. washoensis strain Sb944nv (AF470616), which was isolated from a California ground squirrel (Spermophilus beecheyi) and was genetically identical to an isolate from a human patient with myocarditis (9). The sequence similarities of these genotypes and B. washoensis strain Sb944nv ranged from 94.2% to 97.4%. Genogroup A contained B. washoensis–like genotypes; the genotypes from each squirrel species formed a separate clade, except for the genotypes from Richardson’s ground squirrels (Sp. richardsonii) and Columbian ground squirrels (Sp. columbianus), which formed a mixed clade (Figure).
In this study, 18 genotypes formed the 6 unique genogroups B to G. The DNA sequences of the genotypes in each genogroup showed relatively low similarity (82.4%–94.6%) to the type strains of known Bartonella spp., and sequence similarities between genogroups B to G were also low (87.5%–93.6%). The novel Bartonella genogroups B, C, D, and E were isolated from greater Egyptian jerboas, tricolored squirrels (Callosciurus notatus), fat-tailed gerbils (Pachyuromys duprasi), and golden spiny mice (A. russatus), respectively. The genotypes in group F were isolated from 6 animal species: large Egyptian gerbils, fat-tailed gerbils, fat sand rats (Psammomys obesus), lesser Egyptian jerboas, greater Egyptian jerboas, and bushy-tailed jirds; those in genogroup G were isolated from a bushy-tailed jird and a large Egyptian gerbil (Figure). In genogroup F, 3 of the 9 isolates from fat-tailed gerbils and the 14 isolates from fat sand rats had identical gltA DNA sequences. Furthermore, 5 of the 7 isolates from greater Egyptian jerboas and 2 of the 3 isolates from a lesser Egyptian jerboa also had identical sequences.
The 2 novel genotypes (AB444993 and AB444994) in genogroup H were also isolated from Siberian chipmunks, a Hokkaido squirrel (Sciurus vulgaris subsp. orientis), and Eurasian small flying squirrels (Pteromys volans). Their sequences showed high similarity (98.4%–98.7%) to B. grahami type strain (V2) (Figure).
The 10 novel genotypes in genogroup I were isolated from 9 animal species, and the sequence similarities between the genotypes (AB444995 to AB445005) and B. elizabethae type strain (F9251) ranged from 95.5% to 98.7%. The DNA sequences of gltA of the 3 isolates from a Cairo spiny mouse (AB445000) were identical to that of B. elizabethae (F9251). The sequences of the 13 isolates from lesser Egyptian jerboas were identical to those of the 16 isolates from greater Egyptian jerboas.
In genogroup J, a unique genotype (AB445006) was isolated from an American red squirrel (Tamiasciurus hudosonicus); it had 96.2% sequence similarity to B. clarridgeiae type strain (Houston-2), whose natural reservoir is cats (Figure).
Multiple Infections with Different Bartonella Genogroups and Genotypes
Of the 142 Bartonella-positive animals, 25 (17.6%) were found to be infected with different Bartonella genogroups or genotypes (Table 2). A lesser Egyptian jerboa carried 3 different genotypes in 2 genogroups; the other 24 animals carried 2 different genogroups or genotypes. Of these 24 animals, an American red squirrel carried a B. washoensis–like strain in genogroup A and B. clarridgeiae–like strains in genogroup J; 11 animals were infected with B. elizabethae–like strains in genogroup I and strains in genogroups B, C, D, E, F, or G, and the remaining 12 carried different genotypes in the same genogroup (Table 2).
We report prevalence of bartonellae in exotic small mammals imported into Japan as pets. We found that 26.0% (142/546) of the animals examined had bartonellae in their blood. Prevalence among wild captive animal species was high (37.3%), significantly higher (p<0.001) than that among animals from breeder facilities. Of the 179 animals (representing 9 species) from breeder facilities, only 5 Siberian chipmunks imported from a Chinese breeder were found to carry bartonellae, and these were of the same genotype as bartonellae from wild captive animals. These results suggest that animals in breeder facilities may be maintained under hygienic conditions from birth to export, so they rarely have contact with wild animals or blood-sucking arthropod vectors.
Most isolates from animals in the family Sciuridae (58.7%; 122/208) were in genogroup A and showed high sequence similarity to B. washoensis. Kosoy et al. (9) have reported that B. washoensis is widely distributed in ground squirrels in the western part of the United States and that it was isolated from a human with myocarditis in Nevada, USA. Thus, captive squirrels carrying B. washoensis–like organisms could serve as a source of infection for humans.
Animals in the family Sciuridae were also found to be carrying several genotypes of bartonellae in genogroups C, B. grahamii–like strains in genogroup H, B. elizabethae–like strains in genogroup I, and B. clarridgeiae–like strains in genogroup J. The sequence similarities between the genotypes and the related Bartonella spp. type strains ranged from 98.4% to 98.7% for B. grahamii, from 95.5% to 95.8% for B. elizabethae, and were 96.2% for B. clarridgeiae. In humans, B. grahamii, B. elizabethae, and B. clarridgeiae have been reported to cause neuroretinitis (11), endocarditis (10), and cat-scratch disease (31), respectively. These findings suggest that exotic squirrels also might be a potential source of Bartonella infections in humans. Although B. clarridgeiae has mainly been isolated from cats (1), B. clarridgeiae–like strains were isolated from an American red squirrel in this study. B. clarridgeiae–like organisms have also been isolated from yellow-necked mice (Apodemus flavicollis) in Sweden (14) and Greece (15).
The sequence similarity of the gltA sequence (312 bp) of the B. clarridgeiae–like genotype isolated in our study to that of the strain isolated from the yellow-necked mouse (AF391788) was relatively high (97.7%). Recently, B. rochalimae, a B. clarridgeiae–like organism, was isolated from a human patient with bacteremia, fever, and splenomegaly (32). The B. clarridgeiae–like strain from the American red squirrel in this study also showed high gltA sequence similarity (96.8%) with that of B. rochalimae strain BMGH. Studies will be required to clarify the pathogenicity of B. clarridgeiae–like organisms for humans. Such studies would include 1) evaluation of the organisms’ ability to invade human erythrocytes and/or endothelial cells, 2) demonstration of the presence and expression of the genes of type 4 secretion systems (VirB/VirD4 or Vbh) and Trw, and 3) comparisons of the entire genome sequences of the organisms and with those of other human pathogenic Bartonella spp.
In this study, Bartonella genogroups D, E, and G were isolated from animals in the family Muridae, and Bartonella genogroup B was isolated from animals in the family Dipodidae. These findings suggest strict host specificity between the strains in these genogroups and the host animal family. However, findings also showed wide host species diversity; strains in genogroup F were isolated from 6 animal species, and strains from genogroup I (B. elizabethae–like) were isolated from 9 animal species. Bartonella strains in genogroup F were isolated from animals in the families Muridae and Dipodidae. Genogroup I (B. elizabethae–like) strains were also isolated from animals in the family Sciuridae. B. elizabethae has been isolated from different animal species, e.g., a human patient and genus Rattus rats (5,10), and B. elizabethae DNA has been isolated from a dog (33). In our study, 3 Bartonella isolates from a Cairo spiny mouse imported from Egypt had an identical gltA sequence to that of the B. elizabethae type strain. Thus, some Bartonella spp., such as B. elizabethae and B. washoensis, infect host animals in diverse families and may have zoonotic potential.
In the present study, 17.6% (25/142) of exotic animals were infected with different Bartonella genotypes or genogroups. In particular, 3 isolates from a greater Egyptian jerboa were classified in 3 different genotypes. Of the 25 Bartonella-positive animals, 13 showed co-infection with different Bartonella genogroups. Of these 13 animals, 12 carried B. elizabethae–like strains in genogroup I. In contrast, strains with identical gltA sequences were isolated from 2 different animal species, such as greater Egyptian jerboas and lesser Egyptian jerboas, Siberian chipmunks and Hokkaido squirrels, and fat-tailed gerbils and fat sand rats. These findings suggest that some Bartonella species have a wide host range and may be transmitted horizontally by some blood-sucking arthropod vectors with low host specificity.
In summary, we examined the possibility that exotic small mammals may be reservoirs of zoonotic Bartonella spp. around the world. The animals in this study carried, at high prevalence, several Bartonella spp. that are human pathogens. Novel species were suggested by the fact that some of the genotypes in 6 genogroups (B to G) showed relatively low similarity (<94.6%) to known Bartonella spp. and formed independent clusters according to phylogenetic analysis based on partial gltA sequences. More taxonomic studies should sequence other housekeeping genes, such as rpoB, 16S rRNA, ftsZ, groEL, and ribC, to confirm whether these isolates are novel Bartonella spp. (34). To prevent human infections by Bartonella spp. carried by exotic small mammals, a quarantine system for these animals should be established as early as possible. Further studies will be necessary to clarify the route of transmission among exotic small mammals and to evaluate the pathogenicity for humans and animals of the isolates belonging to novel Bartonella genotypes found in this study.
Dr Inoue is a PhD candidate graduate student in the Department of Veterinary Medicine, College of Bioresource Sciences, Nihon University, Japan, and a research assistant at Nihon University Veterinary Research Center. His primary research interests are the epidemiology, ecology, and prevention of bacterial zoonoses.
Acknowledgment
This work was supported in part by a grant for Academic Frontier Project, Surveillance and Control for Zoonoses, from the Ministry of Education, Culture, Sports, Science and Technology, Japan. It was also supported in part by a grant-in-aid from the Ministry of Health, Labour and Welfare, Japan.
References
- Chomel BB, Boulouis HJ, Maruyama S, Breitschwerdt EB. Bartonella spp. in pets and effect on human health. Emerg Infect Dis. 2006;12:389–94.PubMedGoogle Scholar
- Bermond D, Heller R, Barrat F, Delacour G, Dehio C, Alliot A, Bartonella birtlesii sp. nov., isolated from small mammals (Apodemus spp.). Int J Syst Evol Microbiol. 2000;50:1973–9.PubMedGoogle Scholar
- Birtles RJ, Harrison TG, Saunders NA, Molyneux DH. Proposals to unify the genera Grahamella and Bartonella, with descriptions of Bartonella talpae comb. nov., Bartonella peromysci comb. nov., and three new species, Bartonella grahamii sp. nov., Bartonella taylorii sp. nov., and Bartonella doshiae sp. nov. Int J Syst Bacteriol. 1995;45:1–8.PubMedGoogle Scholar
- Welch DF, Carroll KC, Hofmeister EK, Persing DH, Robison DA, Steigerwalt AG, Isolation of a new subspecies, Bartonella vinsonii subsp. arupensis, from a cattle rancher: identity with isolates found in conjunction with Borrelia burgdorferi and Babesia microti among naturally infected mice. J Clin Microbiol. 1999;37:2598–601.PubMedGoogle Scholar
- Ellis BA, Regnery RL, Beati L, Bacellar F, Rood M, Glass GG, Rats of the genus Rattus reservoir hosts for pathogenic Bartonella species: an Old World origin for a New World disease? J Infect Dis. 1999;180:220–4. DOIPubMedGoogle Scholar
- Heller R, Riegel P, Hansmann Y, Delacour G, Bermond D, Dehio C, Bartonella tribocorum sp. nov., a new Bartonella species isolated from the blood of wild rats. Int J Syst Bacteriol. 1998;48:1333–9.PubMedGoogle Scholar
- Gundi VA, Davoust B, Khamis A, Boni M, Raoult D, La Scola B. Isolation of Bartonella rattimassiliensis sp. nov. and Bartonella phoceensis sp. nov. from European Rattus norvegicus. J Clin Microbiol. 2004;42:3816–8. DOIPubMedGoogle Scholar
- Pretorius AM, Beati L, Birtles RJ. Diversity of bartonellae associated with small mammals inhabiting Free State province, South Africa. Int J Syst Evol Microbiol. 2004;54:1959–67. DOIPubMedGoogle Scholar
- Kosoy M, Murray M, Gilmore RD Jr, Bai Y, Gage KL. Bartonella strains from ground squirrels are identical to Bartonella washoensis isolated from a human patient. J Clin Microbiol. 2003;41:645–50. DOIPubMedGoogle Scholar
- Daly JS, Worthington MG, Brenner DJ, Moss CW, Hollis DG, Weyant RS, Rochalimaea elizabethae sp. nov. isolated from a patient with endocarditis. J Clin Microbiol. 1993;31:872–81.PubMedGoogle Scholar
- Kerkhoff FT, Bergmans AM, van Der Zee A, Rothova A. Demonstration of Bartonella grahamii DNA in ocular fluids of a patient with neuroretinitis. J Clin Microbiol. 1999;37:4034–8.PubMedGoogle Scholar
- Fenollar F, Sire S, Raoult D. Bartonella vinsonii subsp. arupensis as an agent of blood culture–negative endocarditis in a human. J Clin Microbiol. 2005;43:945–7. DOIPubMedGoogle Scholar
- Birtles RJ, Harrison TG, Molyneux DH. Grahamella in small woodland mammals in the U.K.: isolation, prevalence and host specificity. Ann Trop Med Parasitol. 1994;88:317–27.PubMedGoogle Scholar
- Holmberg M, Mills JN, McGill S, Benjamin G, Ellis BA. Bartonella infection in sylvatic small mammals of central Sweden. Epidemiol Infect. 2003;130:149–57. DOIPubMedGoogle Scholar
- Tea A, Alexiou-Daniel S, Papoutsi A, Papa A, Antoniadis A. Bartonella species isolated from rodents, Greece. Emerg Infect Dis. 2004;10:963–4.PubMedGoogle Scholar
- Birtles RJ, Canales J, Ventosilla P, Alvarez E, Guerra H, Llanos-Cuentas A, Survey of Bartonella species infecting intradomicillary animals in the Huayllacallán Valley, Ancash, Peru, a region endemic for human bartonellosis. Am J Trop Med Hyg. 1999;60:799–805.PubMedGoogle Scholar
- Jardine C, Appleyard G, Kosoy MY, McColl D, Chirino-Trejo M, Wobeser G, Rodent-associated Bartonella in Saskatchewan, Canada. Vector Borne Zoonotic Dis. 2005;5:402–9. DOIPubMedGoogle Scholar
- Kosoy MY, Regnery RL, Tzianabos T, Marston EL, Jones DC, Green D, Distribution, diversity, and host specificity of Bartonella in rodents from the Southeastern United States. Am J Trop Med Hyg. 1997;57:578–88.PubMedGoogle Scholar
- Iralu J, Bai Y, Crook L, Tempest B, Simpson G, McKenzie T, Rodent-associated Bartonella febrile illness, southwestern United States. Emerg Infect Dis. 2006;12:1081–6.PubMedGoogle Scholar
- Castle KT, Kosoy M, Lerdthusnee K, Phelan L, Bai Y, Gage KL, Prevalence and diversity of Bartonella in rodents of northern Thailand: a comparison with Bartonella in rodents from southern China. Am J Trop Med Hyg. 2004;70:429–33.PubMedGoogle Scholar
- Winoto IL, Goethert H, Ibrahim IN, Yuniherlina I, Stoops C, Susanti I, Bartonella species in rodents and shrews in the greater Jakarta area. Southeast Asian J Trop Med Public Health. 2005;36:1523–9.PubMedGoogle Scholar
- Ying B, Kosoy MY, Maupin GO, Tsuchiya KR, Gage KL. Genetic and ecologic characteristics of Bartonella communities in rodents in southern China. Am J Trop Med Hyg. 2002;66:622–7.PubMedGoogle Scholar
- Inoue K, Maruyama S, Kabeya H, Yamada N, Ohashi N, Sato Y, Prevalence and genetic diversity of Bartonella species isolated from wild rodents in Japan. Appl Environ Microbiol. 2008;74:5086–92. DOIPubMedGoogle Scholar
- Maruyama S, Nogami S, Inoue I, Namba S, Asanome K, Katsube Y. Isolation of Bartonella henselae from domestic cats in Japan. J Vet Med Sci. 1996;58:81–3.PubMedGoogle Scholar
- Norman AF, Regnery R, Jameson P, Greene C, Krause DC. Differentiation of Bartonella–like isolates at the species level by PCR-restriction fragment length polymorphism in the citrate synthase gene. J Clin Microbiol. 1995;33:1797–803.PubMedGoogle Scholar
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. DOIPubMedGoogle Scholar
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25.PubMedGoogle Scholar
- Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. DOIPubMedGoogle Scholar
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. DOIPubMedGoogle Scholar
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution Int J Org Evolution. 1985;39:783–91. DOIGoogle Scholar
- Kordick DL, Hilyard EJ, Hadfield TL, Wilson KH, Steigerwalt AG, Brenner DJ, Bartonella clarridgeiae, a newly recognized zoonotic pathogen causing inoculation papules, fever, and lymphadenopathy (cat scratch disease). J Clin Microbiol. 1997;35:1813–8.PubMedGoogle Scholar
- Eremeeva ME, Gerns HL, Lydy SL, Goo JS, Ryan ET, Mathew SS, Bacteremia, fever, and splenomegaly caused by a newly recognized Bartonella species. N Engl J Med. 2007;356:2381–7. DOIPubMedGoogle Scholar
- Mexas AM, Hancock SI, Breitschwerdt EB. Bartonella henselae and Bartonella elizabethae as potential canine pathogens. J Clin Microbiol. 2002;40:4670–4. DOIPubMedGoogle Scholar
- La Scola B, Zeaiter Z, Khamis A, Raoult D. Gene-sequence-based criteria for species definition in bacteriology: the Bartonella paradigm. Trends Microbiol. 2003;11:318–21. DOIPubMedGoogle Scholar
Figure
Tables
Cite This ArticleTable of Contents – Volume 15, Number 4—April 2009
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Soichi Maruyama, Department of Veterinary Medicine, College of Bioresource, Nihon University, 1866 Kameino, Fujisawa, Kanagawa 252-8510, Japan
Top