Volume 17, Number 11—November 2011
Dispatch
New Dengue Virus Type 1 Genotype in Colombo, Sri Lanka
Abstract
The number of cases and severity of disease associated with dengue infection in Sri Lanka has been increasing since 1989, when the first epidemic of dengue hemorrhagic fever was recorded. We identified a new dengue virus 1 strain circulating in Sri Lanka that coincided with the 2009 dengue epidemic.
Dengue virus (DENV) is a flavivirus transmitted by Aedes spp. mosquitoes. There are 4 distinct DENV serotypes (DENV-1–4). Infection with a single serotype leads to long-term protective immunity against the homologous serotype but not against other serotypes (1). Globally, dengue is an emerging disease that causes an estimated 50–100 million infections, 500,000 dengue hemorrhagic fever (DHF) cases, and 22,000 deaths annually (2,3).
Epidemiologic and other studies indicate that risk factors for severe dengue include secondary infection with a heterologous serotype, the strain of infecting virus, and age and genetic background of the host. Studies are under way to further explore the role of these factors in severe disease (1,4).
In Sri Lanka, serologically confirmed dengue was first reported in 1962 (5), but although all 4 virus serotypes were present and there were cases of DHF, only since 1989 has DHF been considered endemic to Sri Lanka (5). Dengue was made a reportable disease in Sri Lanka in 1996, and the largest epidemic (35,008 reported cases, 170 cases/100,000 population, and 346 deaths) occurred in 2009 (6). DHF epidemics in 1989 and 2002–2004 were associated with emergence of new clades of DENV-3 (7,8). We report a new DENV-1 genotype introduced to Sri Lanka before the 2009 epidemic.
The study was approved by the Ethical Review Committee of the Faculty of Medicine, University of Colombo, Sri Lanka, and the Institutional Research Board of the International Vaccine Institute, Seoul, South Korea. Serum samples were obtained in 2009 and early 2010 from patients as part of a Pediatric Dengue Vaccine Initiative (PDVI) fever surveillance study in Colombo, Sri Lanka. Samples were originally tested for dengue by reverse transcription PCR at Genetech Research Institute (Colombo, Sri Lanka). A random subset of dengue-positive samples of all 4 serotypes was sent to the Program in Emerging Infectious Diseases Laboratory at Duke–National University of Singapore Graduate Medical School, Singapore, for virus isolation and sequencing.
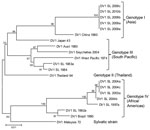
RNA was extracted from virus isolates, subjected to standard reverse transcription PCR to confirm the presence of dengue virus, and serotyped as described (7). Samples processed at Duke–National University of Singapore underwent whole-genome sequencing as described (9). Using DENV-1 isolates from Sri Lanka obtained from dengue cases in 1983, 1984, 1997, 2003, and 2004 (7) and representative DENV-1 sequences for the 4 genotypes, we constructed a phylogenetic tree by using MEGA5 software (10) (Figure; Table).
The 4 DENV serotypes found in Sri Lanka have been classified into genotypes according to the nomenclature described by Rico-Hesse (11). The earliest isolates found in 1983 and 1984 belong to South Pacific genotype III. More recent isolates obtained during surveillance efforts during 1997–2004 belong to Africa/America genotype IV, indicating that at some point between the early 1980s and the mid 1990s, there was a DENV-1 genotype shift. Analysis of viruses isolated in 2009 indicated that another Asia genotype I of DENV-1 has been introduced into Sri Lanka (Figure) (7). This Asia genotype I virus appears to be responsible for the 2009 epidemic of dengue fever and DHF.
A feature of the epidemiology of dengue in Sri Lanka was the lack of DHF in the early 1980s and the increase in the number of severe dengue cases since 1989, more so after 2000. This finding was observed despite seroprevalence rates remaining largely the same over time as reported in a previous study (12) and in the current PDVI study (13).
Previous epidemics (1989 and 2002–2004) showed a correlation with evolution of DENV-3 genotype III in Sri Lanka, where emergence of new clades of DENV-3 genotype 3 showed a correlation with large increases in the number of reported cases and the geographic range of the virus (7,8). A similar observation was reported for Puerto Rico by Bennett et al., who compared data for DEN2 and DEN4 over 20 years and found that dominant clades were replaced by viral subpopulations existing within the population (14) and in the South Pacific region for DENV-2, where a similar clade replacement occurred (15). These clade changes were accompanied by positive selection in the nonstructural protein 2A (NS-2A) gene for DENV-4 and the envelope, premembrane, NS-2A, and NS-4A genes for DENV-2.
Our results indicate that introduction of a new DENV-1 genotype coincided with the 2009 dengue epidemic in Sri Lanka. Studies are underway to determine if the proportion of DENV-1 cases in 2009 was greater than in previous years and to assess the role of this new DENV-1 genotype in the severe epidemic of 2009. Further studies are needed to determine if this new genotype has spread to other countries in the region.
Dr Tissera is a medical epidemiologist at the Epidemiology Unit, Ministry of Health, Colombo, Sri Lanka. His research interests are the epidemiology, virology, surveillance, and control of dengue.
Acknowledgments
We thank the Public Health Department of the Colombo Municipal Council and the Lady Ridgeway Hospital for Children, Colombo, Sri Lanka, for support.
This study was supported by the Bill and Melinda Gates Foundation through the PDVI (grant no. 23197).
References
- Gubler DJ. The global emergence/resurgence of arboviral disease as public health problems. Arch Med Res. 2002;33:330–42. DOIPubMedGoogle Scholar
- World Health Organization. Dengue. Guidelines for diagnosis, treatment, prevention, and control. TDR-WHO [cited 2011 Jul 29]. http://apps.who.int/tdr/svc/publications/training-guideline-publications/dengue-diagnosis-treatment
- Kurane I. Dengue hemorrhagic fever with special emphasis on immunopathogenesis. Comp Immunol Microbiol Infect Dis. 2007;30:329–40. DOIPubMedGoogle Scholar
- Vitarana T, Jayakura WS, Withane N. Historical account of dengue hemorrhagic fever in Sri Lanka. Dengue Bull. 1997;21:117–8.
- Distribution of suspected DF/DHF by week, Sri Lanka 2004–2010. Epidemiology Unit, Ministry of Health, Sri Lanka [cited 2011 Jul 29]. http://www.epid.gov.lk/pdf/dhf-2010/week31-2010-updated-18-08-2010.pdf
- Kanakaratne N, Wahala MP, Messer WB, Tissera HA, Shahani A, Abeysinghe N, Severe dengue epidemics in Sri Lanka, 2003–2006. Emerg Infect Dis. 2009;15:192–9. DOIPubMedGoogle Scholar
- Messer WB, Gubler DG, Harris E, Sivananthan K, de Silva AM. Emergence and global spread of a dengue serotype 3, subtype III virus. Emerg Infect Dis. 2003;9:800–9.PubMedGoogle Scholar
- Christenbury JG, Aw PP, Ong SH, Schreiber MJ, Chow A, Gubler DJ, A method for full genome sequencing of all four serotypes of the dengue virus. J Virol Methods. 2010;169:202–6. DOIPubMedGoogle Scholar
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. DOIPubMedGoogle Scholar
- Rico-Hesse R. Microevolution and virulence of dengue viruses. Adv Virus Res. 2003;59:315–41. DOIPubMedGoogle Scholar
- Messer WB, Vitarana UT, Sivananthan K, Elvtigala J, Preethimala LD, Ramesh R, Epidemiology of dengue in Sri Lanka before and after the emergence of epidemic dengue hemorrhagic fever. [PubMed]. Am J Trop Med Hyg. 2002;66:765–73.PubMedGoogle Scholar
- Tissera HA, De Silva AD, Abeysinghe MR, de Silva AM, Palihawadana P, Gunasena S, Third Vaccine Global Congress, 2009. Dengue surveillance in Colombo, Sri Lanka: baseline seroprevalence among children. Procedia in Vaccinology. 2010;2:109–12 [cited 2011 Sep 29]. http://www.sciencedirect.com/science/article/pii/S1877282X10000214
- Bennett SN, Holmes EC, Chirivella M, Rodriguez DM, Beltran M, Vorndam V, Selection-driven evolution of emergent dengue virus. Mol Biol Evol. 2003;20:1650–8. DOIPubMedGoogle Scholar
- Steel A, Gubler DJ, Bennett SN. Natural attenuation of dengue virus type-2 after a series of island outbreaks: a retrospective phylogenetic study of events in the South Pacific three decades ago. Virology. 2010;405:505–12. DOIPubMedGoogle Scholar
Figure
Table
Cite This ArticleTable of Contents – Volume 17, Number 11—November 2011
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Aruna Dharshan De Silva, Genetech Research Institute, 54 Kitulwatte Rd, Colombo 08, Sri Lanka
Top