Volume 17, Number 11—November 2011
THEME ISSUE
CHOLERA IN HAITI
Dispatch
Toxigenic Vibrio cholerae O1 in Water and Seafood, Haiti
Abstract
During the 2010 cholera outbreak in Haiti, water and seafood samples were collected to detect Vibrio cholerae. The outbreak strain of toxigenic V. cholerae O1 serotype Ogawa was isolated from freshwater and seafood samples. The cholera toxin gene was detected in harbor water samples.
Epidemic cholera is caused by toxigenic strains of Vibrio cholerae serogroups O1 and O139, which spread most often through water contaminated with the bacterium (1). Cholera can also be transmitted by eating contaminated foods, including seafood (2). Like other V. cholerae strains, which are autochthonous in riverine, estuarine, and coastal ecosystems, these strains may persist in the environment (3). An outbreak of cholera began in Haiti’s Artibonite Department in October 2010 and rapidly spread across all 10 Haitian departments and Port-au-Prince. Initial investigations indicated that drinking untreated water was the principal risk factor for infection (4). The ongoing risk posed to the Haitian population through contaminated water raised concern that cholera could also be introduced to other countries through transfer of V. cholerae by ship ballast water, contaminated seafood, or both. To better characterize the contamination of untreated surface water and seafood and to evaluate the risk for V. cholerae transfer from contaminated water in Haitian ports, the US Centers for Disease Control and Prevention (CDC) and the US Food and Drug Administration (FDA) collaborated with the Haitian ministries of health, agriculture, and environment to document the presence of V. cholerae in Haitian freshwater resources and harbors.
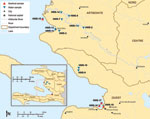
In October and November 2010, water and seafood samples were collected from 2 cholera-affected communities in Haiti and tested for V. cholerae. Eight freshwater and 6 marine water samples were collected from 13 sites in Artibonite and Ouest Departments (Figure). Freshwater samples were collected from rivers, including the Artibonite River, and irrigation canals. Dead-end ultrafiltration, a newly developed technique that has been used to recover diverse microbes from large-volume water samples (5), was used to collect water samples (8–30 L each) at the freshwater sites and 3 of the marine water sites (HWS-11, -13, and -18). At the Haiti National Public Health Laboratory (LNSP), bacteria were recovered from ultrafilters by back flushing with a surfactant solution, and the solution then was added to an equal volume of 2× strength alkaline peptone water (APW). Grab samples (1 L) were collected at 3 harbor sites (HWS-15, -16, and -17) and on arrival at LNSP, they were split into two 500-mL portions for separate testing by CDC at LNSP or for shipment in chilled coolers to FDA (Dauphin Island, AL, USA). At LNSP, all water and ultrafilter back flush samples were incubated in APW at 37°C for 6 h (6). After APW enrichment, the culture broth was streaked onto thiosulfate citrate bile salts (TCBS) agar (Remel, Lenexa, KS, USA) and incubated overnight at 37°C. For each sample, up to 10 colonies suspected of being positive were picked from TCBS agar and grown on nonselective media for multiplex PCR testing (7).
Nine seafood samples were collected along the coast: 5 between Saint-Marc port and Grand Saline (1 site) and 4 from Port-au-Prince port (3 sites) (Figure). Seafood samples were obtained as convenience samples from local fishermen on the water, placed in Ziploc (SC Johnson, Racine, WI, USA) bags, and sent in chilled coolers to FDA, along with 500-mL grab samples of marine water. After enrichment, APW cultures from seafood and water grab samples were tested at FDA by using a real-time PCR specific to the cholera toxin gene (ctxA) of V. cholerae (8). If the APW culture was positive for the ctxA gene, then isolates were obtained by streaking onto TCBS agar as described for water samples.
Identification and characterization of suspected V. cholerae isolates were performed at CDC. Colonies or sweeps of growth typical of V. cholerae were selected from TCBS plates and tested by multiplex PCR for ctxA, tcpAEl Tor, tcpAClassical, ompW, and toxR genes (9–11). Colonies positive by PCR for ctxA or other V. cholerae markers were tested for agglutination in serogroup O1 antiserum and, if positive, also in Inaba and Ogawa serotype antisera. V. cholerae isolates were subtyped according to the PulseNet standardized pulsed-field gel electrophoresis (PFGE) protocol, using primary and secondary restriction enzymes SfiI and NotI, respectively (12). The cholera toxin gene, ctxAB, and tcpA gene regions were amplified by PCR and sequenced (13,14).
V. cholerae O1, serotype Ogawa, ctxA-positive strains were isolated from 2 irrigation canals north of Port-au-Prince in Ouest Department (Table 1). Both of these canals were used for drinking water by the local population, and communities near the canals were heavily affected by the outbreak. V. cholerae O1 Ogawa ctxA-positive strains were isolated from 1 mixed seafood sample (sample 7, containing multiple vertebrate fish and 1 crab) and 1 bivalve sample (sample 8, containing multiple species) that were obtained from fishermen at 2 different locations in the Port-au-Prince port (Table 2). All ctxA-positive V. cholerae isolates were indistinguishable from the outbreak strain by PFGE with both enzymes (pattern combination KZGS12.0088/KZGN11.0092) (15). Sequence analysis for the toxigenic V. cholerae isolates provided additional evidence that the isolates from these samples matched the isolates from humans infected with the outbreak strains. The tcpA sequence of the freshwater and human isolates from Haiti matched that of CIRS 101, an altered El Tor strain from Bangladesh, and the ctxAB sequences matched the sequences from strains isolated in 2007 during an outbreak in Orissa, India (15). The ctxAB and tcpA sequences differed by 1 nt polymorphism from prototypical classical and El Tor alleles, respectively. These isolates were recovered from 30-L freshwater samples having turbidities of 11 and 16 nephelometric turbidity units, which were among those with the lowest turbidity collected during this investigation. All V. cholerae non-O1 ctxA-negative strains possessed unique PFGE patterns distinct from the outbreak pattern. In addition to samples from which toxigenic V. cholerae was isolated, real-time PCR testing by FDA detected the ctxA gene in APW culture broths for 3 seawater samples and 3 other seafood samples.
Isolation of the outbreak strain in seafood samples from Port-au-Prince and detection of the ctxA gene in APW cultures of water and seafood samples from Port-au-Prince and Saint-Marc suggest that harbor waters were contaminated with toxigenic V. cholerae O1. This finding underscores the need for adherence to public health recommendations disseminated during the outbreak regarding making drinking water safe and cooking seafood thoroughly to prevent infection and conducting ship ballast water exchange to limit potential transfer of the organism to other harbors. We report recovery of V. cholerae O1 from large-volume water samples by use of ultrafiltration. Although V. cholerae O1 was not isolated from marine water samples, real-time PCR detection of ctxA in these samples provided additional evidence that harbor water samples were contaminated with toxigenic V. cholerae. Use of this real-time PCR method has provided analytical data that reflected the presence of viable V. cholerae in marine water samples (8). Further assessment by using high-volume filtration and seafood sampling may be useful for tracking the persistence of the strain in the Haitian environment in the future.
Dr Hill is a research environmental engineer at CDC and the team lead for the Water, Sanitation, and Hygiene Laboratory Team in the Division of Foodborne, Waterborne, and Environmental Diseases, Waterborne Disease Prevention Branch. His research interests include the development and application of environmental sampling methods, microbial detection methods, and treatment technologies for water and wastewater systems.
Acknowledgment
We thank the following colleagues for their assistance with this investigation: Valdo Jean, Lucien Revange, Jean Evens Charles, Jocelin Villier, Gabriel Richard, Yves Chartier, and George Roark.
References
- Sasaki S, Suzuki H, Igarashi K, Tambatamba B, Mulenga P. Spatial analysis of risk factor of cholera outbreak for 2003–2004 in a peri-urban area of Lusaka, Zambia. Am J Trop Med Hyg. 2008;79:414–21.PubMedGoogle Scholar
- Finelli L, Swerdlow D, Mertz K, Ragazzoni H, Spitalny K. Outbreak of cholera associated with crab brought from an area with epidemic disease. J Infect Dis. 1992;166:1433–5. DOIPubMedGoogle Scholar
- Colwell RR. Global climate and infectious disease: the cholera paradigm. Science. 1996;274:2025–31. DOIPubMedGoogle Scholar
- O’Connor KA, Cartwright E, Loharikar A, Routh J, Gaines J, Fouché M-DB, Risk factors early in the 2010 cholera epidemic, Haiti. Emerg Infect Dis. 2011;17:2136–8.
- Smith CM, Hill VR. Dead-end hollow-fiber ultrafiltration for recovery of diverse microbes from water. Appl Environ Microbiol. 2009;75:5284–9. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Laboratory methods for the diagnosis of Vibrio cholerae. Atlanta: The Centers; 1994.
- Harris JR, Cavallaro EC, de Nobrega AA, Barrado J, Bopp C, Parsons MB, Field evaluation of Crystal VC Rapid Dipstick test for cholera during a cholera outbreak in Guinea-Bissau. Trop Med Int Health. 2009;14:1117–21. DOIPubMedGoogle Scholar
- Blackstone GM, Nordstrom JL, Bowen MD, Meyer RF, Imbro P, DePaola A. Use of a real time PCR assay for detection of the ctxA gene of Vibrio cholerae in an environmental survey of Mobile Bay. J Microbiol Methods. 2007;68:254–9. DOIPubMedGoogle Scholar
- Nandi B, Nandy RK, Mukhopadhyay S, Nair GB, Shimada T, Ghose AC. Rapid method for species-specific identification of Vibrio cholerae using primers targeted to the gene of outer membrane protein OmpW. J Clin Microbiol. 2000;38:4145–51.PubMedGoogle Scholar
- Keasler SP, Hall RH. Detecting and biotyping Vibrio cholerae O1 with multiplex polymerase chain reaction. Lancet. 1993;341:1661. DOIPubMedGoogle Scholar
- Fields PI, Popovic T, Wachsmuth K, Olsvik O. Use of polymerase chain reaction for detection of toxigenic Vibrio cholerae O1 strains from the Latin American cholera epidemic. J Clin Microbiol. 1992;30:2118–21.PubMedGoogle Scholar
- Cooper KLF, Luey CKY, Bird M, Terajima J, Nair GB, Kam KM, Development and validation of a PulseNet standardized pulsed-field gel electrophoresis protocol for subtyping of Vibrio cholerae. Foodborne Pathog Dis. 2006;3:51–8. DOIPubMedGoogle Scholar
- Li M, Shimada T, Morris JG, Sulakvelidze A, Sozhamannan S. Evidence for the emergence of non-O1 and non-O139 Vibrio cholerae strains with pathogenic potential by exchange of O-antigen biosynthesis regions. Infect Immun. 2002;70:2441–53. DOIPubMedGoogle Scholar
- Boyd EF, Waldor MK. Evolutionary and functional analyses of variants of the toxin-coregulated pilus protein TcpA from toxigenic Vibrio cholerae non-O1/non-O139 serogroup isolates. Microbiology. 2002;148:1655–66.PubMedGoogle Scholar
- Talkington D, Bopp C, Tarr C, Parsons MB, Dahourou G, Freeman M, Characterization of toxigenic Vibrio cholerae from Haiti, 2010–2011. Emerg Infect Dis. 2011;17:2122–9.
Figure
Tables
Cite This ArticleTable of Contents – Volume 17, Number 11—November 2011
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Vincent R. Hill, Centers for Disease Control and Prevention, 1600 Clifton Rd NE, Mailstop D66, Atlanta, GA 30333, USA
Top