Volume 17, Number 2—February 2011
Dispatch
Novel HIV-1 Recombinant Forms in Antenatal Cohort, Montreal, Quebec, Canada
Figure
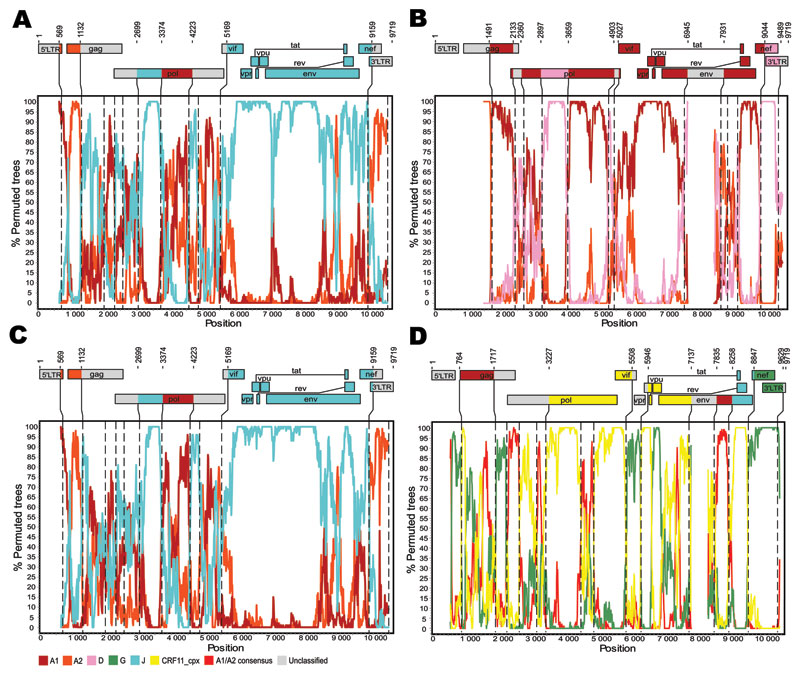
Figure. Genetic organization and recombination breakpoints in HIV-1 genomic sequences isolated from patients TV721 (A), TV725 (B), TV749 (C), and TV919 (D). Nucleotide sequences were submitted to GenBank (accession nos. HM215249–HM215252). Similarity plots were produced with Simplot version 3.5.1 (http://sray.med.som.jhmi.edu/SCRoftware/simplot) by using windows of 500 nt and increments of 50 nt to guide the choice of reference sequences used for bootscanning (8). Bootscan analyses were then performed according to the neighbor-joining method and Kimura 2-parameter distances. The size of the sliding window was set at 300 nt with 10-nt increments (9). Reference sequences used were subtype A1: A1.AU.03 (DQ676872), A1.KE.94 (AF004885), A1.RW.92 (AB253421); subtype A2: A2.CD.97 (AF286238), A2.CY.94 (AF286237); subtype D: D.CD.83.ELI (K03454), D.CM.01 (AY371157), D.TZ.01 (AY253311); subtype G: G.BE.96 (AF084936), G.KE.93 (AF061641), G.NG.92 (U88826); subtype J: J.CD.97 (EF614151), J.SE.93 (AF082394), J.SE.94 (AF082395); and CRF11_cpx: 11_cpx.CM.95 (AF492624). Phylogenetic reconstructions based on the neighbor-joining method and the Kimura 2-parameter distance model were computed by MEGA4 (10) and used to confirm the structures of the recombinants. Bootstrap values >80% (500 replicates) were considered significant. Vertical dashed lines indicate the position of recombination breakpoints. Numbering of residues is based on the sequence of HIV-1 HXB2 (GenBank accession no. K03455).
References
- Leitner T, Foley B, Hahn B, Marx P, McCutchan F, Mellors J, In: Theoretical Biology and Biophysics Group, editors. HIV sequence compendium 2005. Los Alamos (NM): Los Alamos National Laboratory; 2005.
- Brodine SK, Mascola JR, Weiss PJ, Ito SI, Porter KR, Artenstein AW, Detection of diverse HIV-1 genetic subtypes in the USA. Lancet. 1995;346:1198–9. DOIPubMedGoogle Scholar
- Weidle PJ, Ganea CE, Irwin KL, Pieniazed D, McGowan JP, Olivo N, Presence of human immunodeficiency virus (HIV) type 1, group M, non-B subtypes, Bronx, New York: a sentinel site for monitoring HIV genetic diversity in the United States. J Infect Dis. 2000;181:470–5. DOIPubMedGoogle Scholar
- Lin HH, Gaschen BK, Collie M, El-Fishaway M, Chen Z, Korber BT, Genetic characterization of diverse HIV-1 strains in an immigrant population living in New York City. J Acquir Immune Defic Syndr. 2006;41:399–404. DOIPubMedGoogle Scholar
- Ntemgwa M, Toni TD, Brenner BG, Routy JP, Moisi D, Oliveira M, Near full-length genomic analysis of a novel subtype A1/C recombinant HIV type 1 isolate from Canada. AIDS Res Hum Retroviruses. 2008;24:655–9. DOIPubMedGoogle Scholar
- Akouamba BS, Viel J, Charest H, Merindol N, Samson J, Lapointe N, HIV-1 genetic diversity in antenatal cohort, Canada. Emerg Infect Dis. 2005;11:1230–4.PubMedGoogle Scholar
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. DOIPubMedGoogle Scholar
- Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkami SS, Novak NG, Full-length human immunodeficiency virus type 1 genomes from subtype C–infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–60.PubMedGoogle Scholar
- Zhang C, Ding N, Wei JF. Different sliding window sizes and inappropriate subtype references result in discordant mosaic maps and breakpoint locations of HIV-1 CRFs. Infect Genet Evol. 2008;8:693–7. DOIPubMedGoogle Scholar
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–9. DOIPubMedGoogle Scholar
- Liu TF, Shafer RW. Web resources for HIV type 1 genotypic-resistance test interpretation. Clin Infect Dis. 2006;42:1608–18. DOIPubMedGoogle Scholar
- Novitsky VA, Gaolekwe S, McLane MF, Ndung’u TP, Foley BT, Vannberg F, HIV type 1 A/J recombinant with a pronounced pol gene mosaicism. AIDS Res Hum Retroviruses. 2000;16:1015–20. DOIPubMedGoogle Scholar
- Saunders-Buell E, Saad MD, Abed AM, Bose M, Todd CS, Strathdee SA, A nascent HIV type 1 epidemic among injecting drug users in Kabul, Afghanistan is dominated by complex AD recombinant strain, CRF35_AD. AIDS Res Hum Retroviruses. 2007;23:834–9. DOIPubMedGoogle Scholar
- Wilbe K, Casper C, Albert J, Leitner T. Identification of two CRF11-cpx genomes and two preliminary representatives of a new circulating recombinant form (CRF13-cpx) of HIV type 1 in Cameroon. AIDS Res Hum Retroviruses. 2002;18:849–56. DOIPubMedGoogle Scholar
- Kiwanuka N, Laeyendecker O, Robb M, Kigozi G, Arroyo M, McCutchan F, Effect of human immunodeficiency virus type 1 (HIV-1) subtype on disease progression in persons from Rakai, Uganda, with incident HIV-1 infection. J Infect Dis. 2008;197:707–13. DOIPubMedGoogle Scholar