Volume 18, Number 2—February 2012
Dispatch
Oseltamivir-Resistant Pandemic (H1N1) 2009 Virus Infections, United States, 2010–11
Abstract
During October 2010–July 2011, 1.0% of pandemic (H1N1) 2009 viruses in the United States were oseltamivir resistant, compared with 0.5% during the 2009–10 influenza season. Of resistant viruses from 2010–11 and 2009–10, 26% and 89%, respectively, were from persons exposed to oseltamivir before specimen collection. Findings suggest limited community transmission of oseltamivir-resistant virus.
During the 2009–2010 influenza pandemic, when pandemic (H1N1) 2009 virus was the predominant circulating influenza virus (1), the prevalence of oseltamivir-resistant pandemic (H1N1) 2009 viruses in the United States was 0.5%. Of the patients with oseltamivir-resistant virus infection, 89% had been exposed to oseltamivir before specimen collection (2). We describe patients with oseltamivir-resistant pandemic (H1N1) 2009 virus infections during the 2010–11 influenza season.
During October 1, 2010–July 31, 2011, the Centers for Disease Control and Prevention (CDC; Atlanta, Georgia, USA) asked all state public health laboratories to submit pandemic (H1N1) 2009 virus–positive respiratory specimens and virus isolates for antiviral susceptibility testing. Laboratories were asked to provide the first 5 specimens of any type/subtype collected during each 2-week period for virus isolation. Comprehensive antiviral testing, including neuraminidase inhibition (NI) assay, was performed on all isolates, and sequencing was performed on all isolates with elevated 50% inhibitory concentration values. CDC also requested that laboratories provide 5 additional specimens every 2 weeks for pyrosequencing to identify the H275Y substitution in the neuraminidase, a change associated with oseltamivir resistance (3). In addition to (or instead of) participating in the national surveillance, state laboratories in California, Maine, Maryland, Minnesota, New York, Texas, and Washington performed pyrosequencing on state surveillance specimens to detect the H275Y substitution. We included those state-specific data in the national surveillance data for this report. State health departments used a standard case form to collect demographic and clinical information for patients with oseltamivir-resistant pandemic (H1N1) 2009 virus infection and their ill close contacts.
Oseltamivir resistance was determined by use of an NI assay or pyrosequencing for the H275Y substitution. For NI testing on isolates, we used the NA-Fluor kit (Applied Biosystems, Foster City, CA, USA) as described (4). We performed pyrosequencing, as described (5), on all oseltamivir-resistant pandemic (H1N1) 2009 isolates identified by NI assay to confirm the presence of the H275Y substitution. We performed pyrosequencing for H275Y, without the NI assay, to screen pandemic (H1N1) 2009 clinical specimens (5). For the national surveillance, NI testing was performed at CDC and pyrosequencing was performed at CDC and at state laboratories in Wisconsin, New York, and California. State laboratories followed pyrosequencing protocols provided by CDC; when possible, CDC confirmed results for resistant viruses by use of pyrosequencing and NI assay. Most oseltamivir-resistant viruses in this report were included in the weekly FluView report prepared by CDC (6).
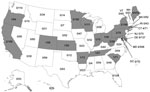
We tested a total of 3,652 pandemic (H1N1) 2009 virus isolates and specimens from every state and the District of Columbia; 35 (1.0%) isolates/specimens from a total of 18 states were oseltamivir-resistant (Figure). Overall, 8 (1.3%) of 609 isolates tested by NI assay and 27 (1.0%) of 3,043 specimens tested by pyrosequencing were resistant to oseltamivir. The state-specific prevalence of oseltamivir-resistant pandemic (H1N1) 2009 viruses varied; however, the number of viruses and specimens tested also varied markedly between states, and several states submitted only a few specimens. Forty-four states submitted >20 specimens for antiviral resistance surveillance. The prevalence of oseltamivir resistance among these specimens ranged from 0% to 5.6%. None of the 609 pandemic (H1N1) 2009 isolates tested by NI assay were resistant to zanamivir. The ranges of 50% inhibitory concentration values for oseltamivir-resistant and -susceptible isolates were 166.17–230.37 nmol/L and 0.10–0.80 nmol/L, respectively.
The median age of the 35 patients with oseltamivir-resistant pandemic (H1N1) 2009 virus infections was 33 years. Of 34 patients with available information, 26% reported receiving oseltamivir before providing a specimen for antiviral susceptibility testing (Table 1). Among 33 patients with a completed case form, 67% had at least 1 preexisting chronic medical condition, 24% had an immunocompromising medical condition, 42% required hospitalization, and 9% died. Most patients with oseltamivir-resistant virus infection for whom housing information was available lived in a single-family household. Two siblings from 1 household had oseltamivir-resistant virus infection; neither child had received oseltamivir.
All oseltamivir-resistant pandemic (H1N1) 2009 viruses were identified from specimens collected during December 2010–April 2011; the prevalence of resistance did not change significantly over time (test for trend, p = 0.18) (Table 2). In addition, the proportion of patients with oseltamivir-resistant virus infections who did not have exposure to oseltamivir before specimen collection did not change significantly over time (test for trend, p = 0.48); however, the number of specimens tested each month was small.
The number of oseltamivir-resistant pandemic (H1N1) 2009 virus–infected patients was small during the 2009–10 (April 2009–September 2010) and 2010–11 influenza seasons. However, the prevalence of oseltamivir-resistant virus–infected patients was slightly higher during 2010–11 compared with 2009–10 (35/3,652 [1.0%] vs. 37/6,740 [0.5%], respectively, p = 0.02 by χ2 test) (2). Also, during 2010–11, compared with 2009–10, more patients with oseltamivir-resistant virus infection had no history of oseltamivir exposure before specimen collection (25/34 [73.5%] vs. 4/35 [11.4%], respectively; p<0.0001 by χ2 test).
During the 2010–11 US influenza season, the prevalence of oseltamivir-resistant pandemic (H1N1) 2009 viruses remained low, and most persons with oseltamivir-resistant virus infection had no prior oseltamivir exposure. This is a notable difference from surveillance findings both globally and in the United States during the 2009–10 season, when most patients with oseltamivir-resistant pandemic (H1N1) 2009 virus infection had a history of oseltamivir exposure, and many were severely immunocompromised, a condition that may increase the risk for resistance developing during oseltamivir therapy (2,7). These data suggest a low level of community transmission of oseltamivir-resistant pandemic (H1N1) 2009 virus in the United States during the 2010–11 influenza season. The United Kingdom also reported that the proportion of oseltamivir-resistant pandemic (H1N1) 2009 virus–infected patients without prior oseltamivir exposure was higher during 2010–11 than 2009–10 (8).
The increase during the 2010-11 influenza season in the proportion of patients with oseltamivir-resistant pandemic (H1N1) 2009 virus infections without prior oseltamivir exposure causes concern in light of the recent history of oseltamivir resistance among seasonal influenza A (H1N1) viruses that circulated before pandemic (H1N1) 2009 virus emerged. In the United States before the 2007–08 influenza season, the prevalence of oseltamivir resistance among seasonal influenza A (H1N1) viruses was <1% (9,10). However, during the 2007–08 season, the prevalence of oseltamivir resistance among seasonal influenza A (H1N1) viruses increased to 12%, and by the 2008–09 season, resistance dramatically increased to >99% (9,11,12). No association was found between this increase in oseltamivir resistance and prior oseltamivir use (11,13). Oseltamivir resistance in pandemic (H1N1) 2009 and seasonal influenza A (H1N1) viruses was conferred by the H275Y substitution in the neuraminidase. However, unlike seasonal influenza A (H1N1) viruses, which retained susceptibility to the M2-blocking adamantanes (amantadine and rimantadine), >99% of circulating pandemic (H1N1) 2009 viruses are inherently resistant to adamantanes (14). Thus, inhaled zanamivir or investigational drugs, such as intravenous zanamivir, are the only treatment options for patients with oseltamivir-resistant pandemic (H1N1) 2009 virus infection.
Our conclusions are limited by the small number of patients with oseltamivir-resistant pandemic (H1N1) 2009 virus infection. Variability in state surveillance and the number of specimens tested from each state may also have limited the representativeness of our data. Despite these shortcomings, our findings emphasize the importance of ongoing surveillance for oseltamivir-resistant pandemic (H1N1) 2009 viruses in the United States and globally and of close monitoring for changes in the epidemiology of oseltamivir resistance among pandemic (H1N1) 2009 viruses. Updated information about antiviral resistance in influenza viruses in the United States is available at www.cdc.gov/flu/professionals/.
Dr Storms is an Epidemic Intelligence Service Officer assigned to the Influenza Division at the Centers for Disease Control and Prevention. His primary research interests are influenza and tropical infectious diseases.
Acknowledgments
Additional members of the US Antiviral Resistance Surveillance Working Group who contributed data: Linda Gladden (Arkansas Department of Health); Janice Louie (California Department of Public Health); Janell Kenfield (Colorado Department of Public Health and Environment); Julie Alan Siniscalchi (State of Connecticut Department of Public Health); Paula Eggers and Emily Outten (Delaware Health and Social Services); Shawn Richards (Indiana State Department of Health); Jena Callen (Kansas Department of Public Health and Environment); Laura Shepherd and Lori Webber (Maine Department of Health and Human Service); René Najera (Maryland Department of Health and Mental Hygiene); Karen Martin (Minnesota Department of Health); Jennifer Lapplante (New York State Department of Health); Matthew Laidler (Oregon Health Authority); Owen Simwale and Kumar Nalluswami (Pennsylvania Department of Health); Brittanni Wright (South Carolina Department of Health and Environmental Control); Robb L. Garman (Tennessee Department of Health); Crystal Van Cleave (Texas Department of State Health Services); Julie Tseng-Crank (Washington State Department of Health); Julie Freshwater (West Virginia Bureau of Public Health); Mary Wedig and Thomas Haupt (Wisconsin Department of Health Services); Shikha Garg, Marnie Levine, Katrina Sleeman, Kristina Ballenger, Angie Trujillo, and Ha Nguyen (Centers for Disease Control and Prevention).
We thank all patients and health care providers who participated in these surveillance activities, as well as state and local public health officials and laboratory staff who assisted in compiling data for this report.
References
- Centers for Disease Control and Prevention. Update: influenza activity—United States, 2009–10 season. MMWR Morb Mortal Wkly Rep. 2010;59:901–8.PubMedGoogle Scholar
- Graitcer SB, Gubareva L, Kamimoto L, Doshi S, Vandermeer M, Louie J, Characteristics of patients with oseltamivir-resistant pandemic (H1N1) 2009, United States. Emerg Infect Dis. 2011;17:255–7.PubMedGoogle Scholar
- Ives JAL, Carr JA, Mendel DB, Tai CY, Lambkin R, Kelly L, The H274Y mutation in the influenza A/H1N1 neuraminidase active site following oseltamivir phosphate treatment leave virus severely compromised both in vitro and in vivo. Antiviral Res. 2002;55:307–17. DOIPubMedGoogle Scholar
- Nguyen HT, Sheu TG, Mishin VP, Klimov AI, Gubareva LV. Assessment of pandemic and seasonal influenza A (H1N1) virus susceptibility to neuraminidase inhibitors in three enzyme activity inhibition assays. Antimicrob Agents Chemother. 2010;54:3671–7. DOIPubMedGoogle Scholar
- Deyde VM, Sheu TG, Trujillo AA, Okomo-Adhiambo M, Garten R, Klimov AI, Detection of molecular markers of drug resistance in 2009 pandemic influenza A (H1N1) viruses by pyrosequencing. Antimicrob Agents Chemother. 2010;54:1102–10. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. FluView. Flu activity and surveillance [cited 2011 Jun 10]. http://www.cdc.gov/flu/weekly/pastreports.htm
- Calatayud L, Lackenby A, Reynolds A, McMenamin J, Phin NF, Zambon M, Oseltamivir-resistant pandemic (H1N1) 2009 virus infection in England and Scotland, 2009-2010. Emerg Infect Dis. 2011;17:1807–15.PubMedGoogle Scholar
- Lackenby A, Moran Gilad J, Pebody R, Miah S, Calatayud L, Bolotin S, Continued emergence and changing epidemiology of oseltamivir-resistant influenza A(H1N1)2009 virus, United Kingdom, winter 2010/11. Euro Surveill. 2011;16;pii:19784. Erratum in Euro Surveill. 2011;16:pii:19789.PubMedGoogle Scholar
- Sheu TG, Deyde VM, Okomo-Adhiambo M, Garten RJ, Xu X, Bright RA, Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob Agents Chemother. 2008;52:3284–92. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Influenza activity—United States and worldwide, 2007–08 season. MMWR Morb Mortal Wkly Rep. 2008;57:692–7.PubMedGoogle Scholar
- Dharan NJ, Gubareva LV, Meyer JJ, Okomo-Adhiambo M, McClinton RC, Marshall SA, Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA. 2009;301:1034–41. DOIPubMedGoogle Scholar
- Centers for Disease Control and Prevention. Update: Influenza activity—United States, September 28, 2008–January 31, 2009. MMWR Morb Mortal Wkly Rep. 2009;58:115–9.PubMedGoogle Scholar
- Meijer A, Lackenby A, Hungnes O, Lina B, van der Werf S, Schweiger B, Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007–08 season. Emerg Infect Dis. 2009;15:552–60. DOIPubMedGoogle Scholar
- Gubareva LV, Trujillo AA, Okomo-Adhiambo M, Mishin VP, Deyde VM, Sleeman K, Comprehensive assessment of 2009 pandemic influenza A (H1N1) virus drug susceptibility in vitro. Antivir Ther. 2010;15:1151–9. DOIPubMedGoogle Scholar
Figure
Tables
Cite This Article1Additional members of the US Antiviral Resistance Surveillance Working Group who contributed data are listed at the end of this article.
Table of Contents – Volume 18, Number 2—February 2012
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Alicia M. Fry, Centers for Disease Control and Prevention, 1600 Clifton Rd NE, Mailstop A34, Atlanta, GA 30333, USA
Top