Volume 19, Number 1—January 2013
Dispatch
Hepatitis E Virus Genotype 4 Outbreak, Italy, 2011
Figure 2
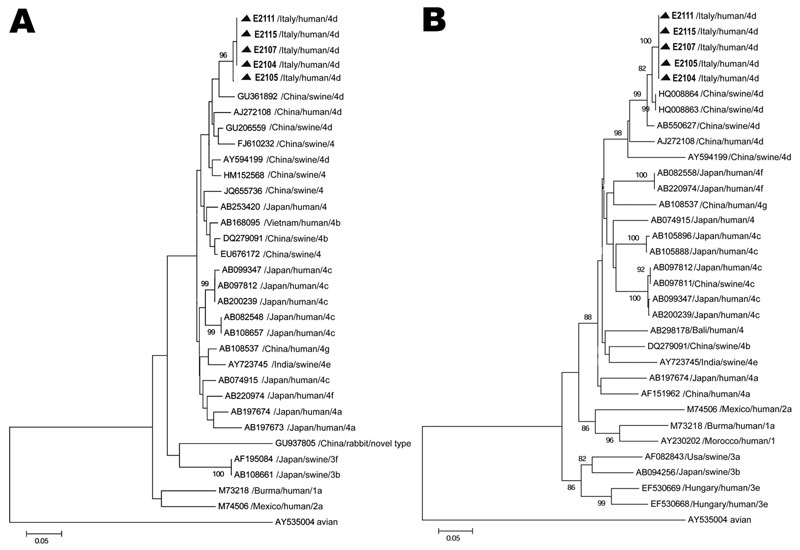
Figure 2. . . Phylogenetic trees based on partial open reading frame (ORF) sequences of the hepatitis E virus monophyletic strain involved in an outbreak in Lazio, Italy, March–April 2011. A) ORF1, 172 nt. Sequences from the outbreak in Italy could not be submitted to GenBank, being <200 nt long; they are available on request from the authors. B) ORF2, 411 nt. The ORF 2 sequence (identical in all 5 patients) described in this panel was submitted to GenBank (accession no. JX401928). Neighbor-joining trees were built by using MEGA5.1 software (www.megasoftware.net), applying the Jukes-Cantor p-distance model of nucleotide substitution. Bootstrap values were determined on 1,000 resamplings of the data set; bootstrap values >80 are shown. Reference strains from GenBank are also included in the trees. Reference viral strains are identified by GenBank accession number, source, country of origin, and respective genotype and subtype. The avian strain AY535004 was used as outgroup. Triangles indicate sequences recovered during the outbreak in Italy. Scale bars represent nucleotide substitutions per site.