Volume 19, Number 1—January 2013
Dispatch
Characterization of Full Genome of Rat Hepatitis E Virus Strain from Vietnam
Figure
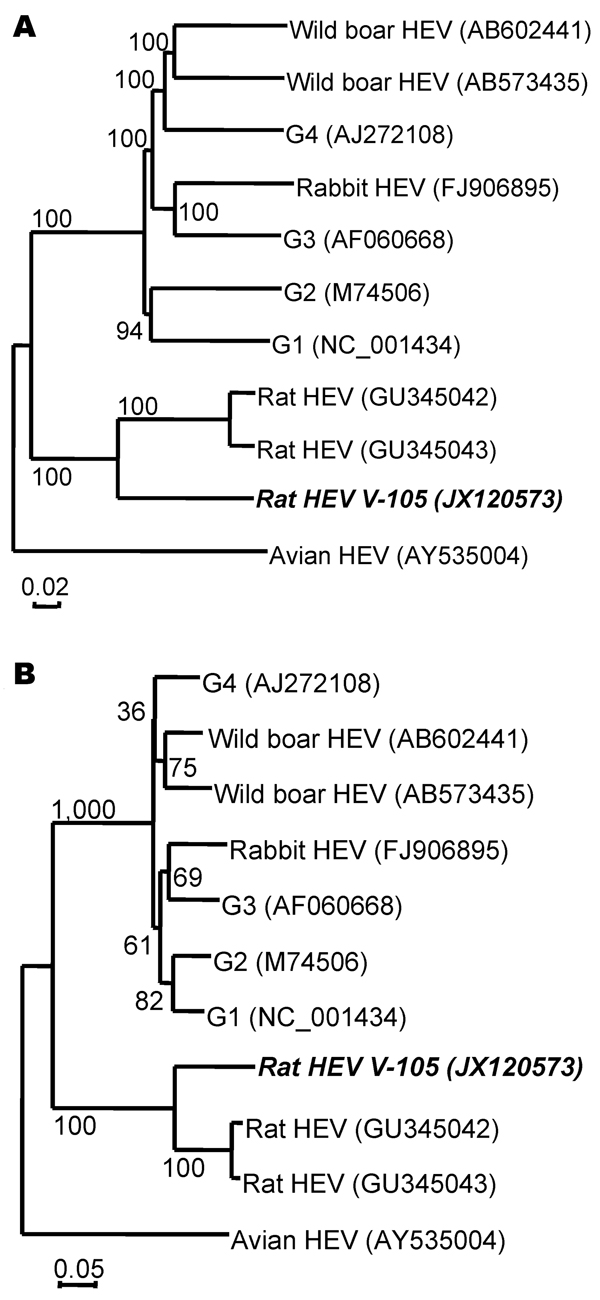
Figure. . Phylogenetic relationships among genotypes 1–4, wild boar, rabbit, avian (chicken), and rat hepatitis E virus (HEV) isolates. The nucleic acid sequence alignment was performed by using ClustalX 1.81 (www.clustal.org). The genetic distance was calculated by the Kimura 2-parameter method. A phylogenetic tree with 1,000 bootstrap replicates was generated by the neighbor-joining method, based on the entire genome (A) and open reading frame 3 (B) of the genotypes 1–4, wild boar, rabbit, avian (chicken), and rat HEV isolates. Scale bar indicates nucleotide substitutions per site. Boldface indicates isolates used in this study.
Page created: January 04, 2013
Page updated: January 04, 2013
Page reviewed: January 04, 2013
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.