Volume 20, Number 11—November 2014
Research
Novel Chlamydia trachomatis Strains in Heterosexual Sex Partners, Indianapolis, Indiana, USA
Figure 2
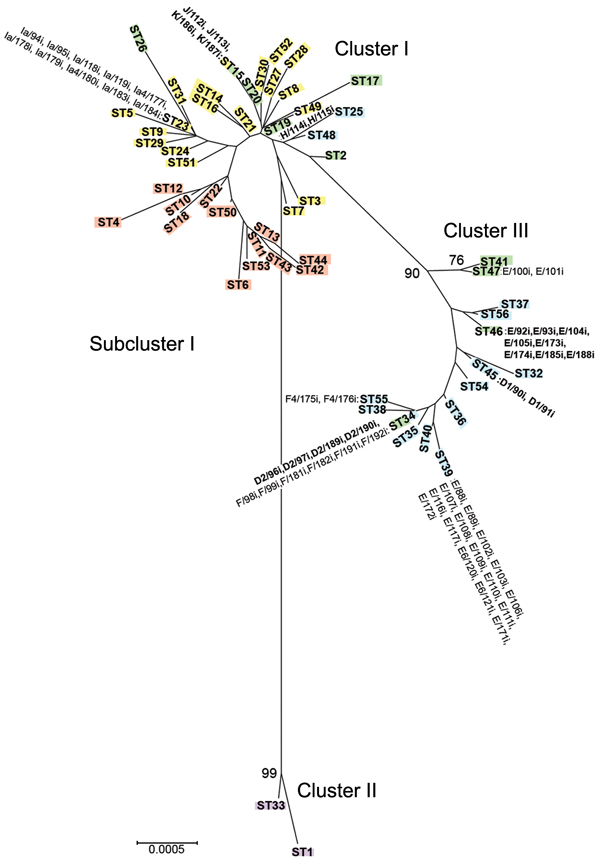
Figure 2. Minimum evolution tree of Chlamydia trachomatis samples collected during April 2000–October 2003 from members of heterosexual partnerships (dyads) in Indianapolis, Indiana, USA, compared with reference strains. The tree was constructed by using the 192 concatenated sequences in the MLST database (http://www.mlst.net) for the 7 loci. Bootstrap values (1,000 replicates) >70% are shown. Three clusters and 1 subcluster are shown: cluster I, yellow, noninvasive, nonprevalent sexually transmitted infection (STI) strains; subcluster I, red, trachoma strains; cluster II, purple, invasive lymphogranuloma venereum strains; and cluster III, blue, noninvasive, highly prevalent STI strains. Green denotes putative recombinant stains. Samples from Indianapolis are indicated next to sequence types; those in boldface are putative recombinants. Scale bar indicates number of substitutions per site.
1These authors contributed equally to this article.