Volume 20, Number 12—December 2014
Dispatch
Two Anaplasma phagocytophilum Strains in Ixodes scapularis Ticks, Canada
Abstract
We developed PCR-based assays to distinguish a human pathogenic strain of Anaplasma phagocytophilum, Ap-ha, from Ap-variant 1, a strain not associated with human infection. The assays were validated on A. phagocytophilum-infected blacklegged ticks (Ixodes scapularis) collected in Canada. The relative prevalence of these 2 strains in I. scapularis ticks differed among geographic regions.
The gram-negative bacterium Anaplasma phagocytophilum is the causative agent of human granulocytic anaplasmosis (HGA) in the United States (1). More than 90% of HGA cases occur in the Upper Midwest and Northeast (2). In these regions, blacklegged ticks (Ixodes scapularis) are the vectors of a human pathogenic strain (Ap-ha) and a variant strain (Ap-variant 1) of A. phagocytophilum (3–7), the latter of which appears not to be associated with human infection (1,3). HGA represents an emerging disease in southern Canada because populations of I. scapularis ticks have become established or are in the process of becoming established (8,9). However, there is limited information on the occurrence of A. phagocytophilum in these ticks (10–12) and the proportion of ticks infected with the Ap-ha strain.
The most commonly used method to distinguish the human pathogenic strain of A. phagocytophilum from those not associated with human infection is to sequence the 16S rRNA gene (3–7,13). The Ap-ha strain differs from the Ap-variant 1 strain by 2 nucleotide differences at the 5′ end of the gene sequence (1,3). However, there is a need for an alternative to currently used PCR-based methods for strain identification that are reliable, but faster and more cost-effective. Therefore, the objectives of the current study were to determine the proportion of blacklegged ticks infected with A. phagocytophilum in different geographic regions of Canada, and to develop restriction fragment length polymorphism (RFLP) and single nucleotide polymorphism (SNP) genotyping assays, targeting the 16S rRNA gene, to differentiate the Ap-ha strain from the Ap-variant 1 strain. We assessed the usefulness of these assays and determined the prevalence of the Ap-ha strain in I. scapularis ticks from different geographic regions of Canada.
We conducted real-time PCR analyses targeting the msp2 gene on genomic (g) DNA of 12,606 I. scapularis ticks collected across Canada (Technical Appendix 1). Of these, 169 (1.3%) ticks were PCR-positive (Table 1). There were significant differences (χ25 = 129.7, p<0.001) in the proportions of ticks infected with A. phagocytophilum among geographic regions; with a greater proportion of PCR-positive ticks in Manitoba than in Ontario, Quebec, and the Atlantic provinces.
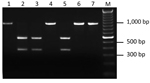
We developed PCR-based assays to distinguish the Ap-ha strain from the Ap-variant 1 strain of A. phagocytophilum based on DNA sequence comparisons of the 16S rRNA gene of each strain (Technical Appendix 1) over a much larger region (875 bp) than in previous studies (1,3). In addition to the 2 nucleotide differences described previously (1,3), a third difference (at position 536) was detected in the aligned sequences (Technical Appendix 2). This nucleotide alteration in the DNA sequence of the Ap-variant 1 strain was associated with a restriction site for the endonuclease Kpn2I (T/CCGGA) that was absent in the sequence of the Ap-ha strain. Two PCR-RFLP assays (Technical Appendix 1), developed at different laboratories, were designed on the basis of this sequence difference and tested on 22 amplicons derived from A. phagocytophilum–infected ticks collected in Minnesota, USA (n = 17) and Manitoba (n = 5). Identical results were obtained for both assays. Three different RFLP patterns were produced. Eighteen amplicons remained undigested (i.e., a single band of ≈920 bp), and 3 amplicons had 2 bands (≈360 and ≈550 bp), representing the expected patterns for the Ap-ha strain and Ap-variant 1 strain, respectively (Figure 1). These results were confirmed on the basis of comparisons of the DNA sequences of representative samples. The RFLP pattern of 1 amplicon, derived from the gDNA of a female tick from Itasca State Park, Minnesota, consisted of 3 bands (≈360, ≈550 and ≈920 bp) (Technical Appendix 2), suggesting a mixture of the 2 strains. Additional analyses of another 125 amplicons from A. phagocytophilum-infected ticks revealed that 79 (63%) had RFLP patterns consistent with the Ap-variant 1 strain, and 46 (37%) had RFLP patterns of the Ap-ha strain. The DNA sequences of a subset of these samples (n = 58) showed 100% agreement between RFLP pattern and strain type of A. phagocytophilum (i.e., 24 of the Ap-ha strain and 34 of the Ap-variant 1 strain).
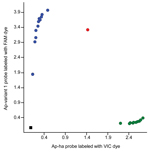
We also designed a custom TaqMan SNP genotyping assay (https://www.lifetechnologies.com/order/custom-genomic-products/tools/genotyping) to differentiate the two A. phagocytophilum strains on the basis of a nucleotide difference at the 5′ end of the 16S rRNA gene sequence (Technical Appendix 1). The SNP assay clearly discriminated between ticks infected with the Ap-ha and/or Ap-variant 1 strains (Figure 2). Of the 142 amplicons tested by this assay, 82 (58%) contained the Ap-variant 1 strain, 59 (42%) contained the Ap-ha strain, and 1 contained a mixture of both strains, which was in 100% agreement with the results of the RFLP analyses and DNA sequencing. The results of the SNP analyses also revealed a significant difference (χ22 = 40.48, p<0.001) in the proportions of I. scapularis ticks infected with the Ap-ha strain among geographic regions (Table 2). A smaller proportion of ticks from Central Provinces were infected with the Ap-ha strain when compared with those from the Prairie and Atlantic Provinces.
A small proportion (1.3%) of I. scapularis ticks collected in Canada were infected with A. phagocytophilum. The prevalence of A. phagocytophilum–-infected ticks differed among geographic regions, but the potential significance of this finding needs to be explored further. Although knowledge of the prevalence of A. phagocytophilum–infected I. scapularis ticks provides some information for determining the public health risks for HGA, the prevalence of the Ap-ha strain of A. phagocytophilum in blacklegged ticks should be considered for risk assessment, since the Ap-ha strain, and not the Ap-variant 1 strain, is most often associated with clinical cases of HGA (4).
For the current study, 3 PCR-based assays were developed to distinguish the Ap-ha strain from the Ap-variant 1 strain. DNA sequencing of representative samples confirmed the reliability of these assays. Each of the 3 assays detected the presence of both strains in the gDNA of a female tick. Mixed infections of both strains in I. scapularis ticks have been reported (4), but appear to be uncommon. Results of these assays also were 100% concordant with the results of 2 RFLP assays (developed at different laboratories) that were tested on the same gDNA samples. Similarly, there was total concordance in the identifications of the A. phagocytophilum strains present within 125 infected ticks collected across Canada by using the seminested PCR-RFLP assay and the TaqMan SNP genotyping assay. The results of these analyses revealed significant differences in the proportion of blacklegged ticks infected with the Ap-ha strain among geographic regions (p<0.001). The public health implications of these findings need to be examined in more detail, using the molecular assays developed in this study.
The TaqMan SNP genotyping assay is ideal for clinical and epidemiologic use where it may be essential to distinguish between the 2 strains of A. phagocytophilum in I. scapularis to assess the potential risk for human infection. However, in a clinical setting, it remains to be established how this assay would be incorporated into or supplement the current diagnostic approach for detecting A. phagocytophilum infections in humans. This test is less technically demanding and takes less time to perform than nested/seminested PCR-RFLPs and DNA sequencing analyses. Moreover, this method eliminates the need for postamplification manipulations and technical problems that are sometimes associated with RFLP analyses of amplicons produced by nested PCR (14,15). Nonetheless, given the 100% concordance in the results of the different analytical methods, the PCR-RFLP assays provide a reliable and cost-effective approach for distinguishing the Ap-ha strain from the Ap-variant 1 strain of A. phagocytophilum. The PCR-RFLP assays will be particularly useful in research laboratories that lack the capacity to conduct real-time PCRs providing an independent and relatively inexpensive method to confirm the results of the SNP assay.
Ms Krakowetz is a PhD candidate in the Biology Department at the University of Saskatchewan. Her research focus is on the population genetics and bacteria of blacklegged ticks in Canada.
Acknowledgments
We thank David Nietzel and Melissa Kemperman (Minnesota Department of Health) for providing the blacklegged ticks from Minnesota, and Tim Kurtti (University of Minnesota) for supplying the equine isolate of A. phagocytophilum [MN-93]. We also thank Shaun Dergousoff and Allison Sproat for laboratory assistance.
Funding for this work was provided by the Natural Sciences and Engineering Research Council of Canada (Discovery Grant to N.B.C., CGSM and Vanier Canada Graduate Scholarships to C.N.K.), the Canadian Foundation for Innovation (to N.B.C.), and the Provincial Government of Saskatchewan and the University of Saskatchewan (Saskatchewan Innovation and Opportunity Scholarship (to C.N.K.).
References
- Chen SM, Dumler JS, Bakken JS, Walker DH. Identification of a granulocytotropic Ehrlichia species as the etiologic agent of human disease. J Clin Microbiol. 1994;32:589–95 .PubMedGoogle Scholar
- Dahlgren FS, Mandel EJ, Krebs JW, Massung RF, McQuiston JH. Increasing incidence of Ehrlichia chaffeensis and Anaplasma phagocytophilum in the United States, 2000–2007. Am J Trop Med Hyg. 2011;85:124–31 . DOIPubMedGoogle Scholar
- Massung RF, Slater K, Owens JH, Nicholson WL, Mather TN, Solberg VB, Nested PCR assay for detection of granulocytic ehrlichiae. J Clin Microbiol. 1998;36:1090–5 .PubMedGoogle Scholar
- Massung RF, Mauel MJ, Owens JH, Allan N, Courtney JW, Stafford KC, Genetic variants of Ehrlichia phagocytophila, Rhode Island and Connecticut. Emerg Infect Dis. 2002;8:467–72. DOIPubMedGoogle Scholar
- Courtney JW, Dryden RL, Wyleto P, Schneider BS, Massung RF. Characterization of Anaplasma phagocytophila and Borrelia burgdorferi genotypes in Ixodes scapularis ticks from Pennsylvania. Ann N Y Acad Sci. 2003;990:131–3. DOIPubMedGoogle Scholar
- Michalski M, Rosenfield C, Erickson M, Selle R, Bates K, Essar D, Anaplasma phagocytophilum in central and western Wisconsin: a molecular survey. Parasitol Res. 2006;99:694–9. DOIPubMedGoogle Scholar
- Steiner FE, Pinger RR, Vann CN, Grindle N, Civitello D, Clay K, Infection and co-infection rates of Anaplasma phagocytophilum variants, Babesia spp., Borrelia burgdorferi, and the rickettsial endosymbiont in Ixodes scapularis (Acari: Ixodidae) from sites in Indiana, Maine, Pennsylvania, and Wisconsin. J Med Entomol. 2008;45:289–97. DOIPubMedGoogle Scholar
- Ogden NH, Lindsay LR, Morshed M, Sockett PN, Artsob H. The emergence of Lyme disease in Canada. CMAJ. 2009;180:1221–4 and. DOIPubMedGoogle Scholar
- Bouchard C, Beauchamp G, Nguon S, Trudel L, Milord LF, Lindsay LR, Associations between Ixodes scapularis ticks and small mammal hosts in a newly endemic zone in southeastern Canada: implications for Borrelia burgdorferi transmission. Ticks Tick Borne Dis. 2011;2:183–90.
- Ogden NH, Lindsay RL, Hanincova K, Barker IK, Bigras-Poulin M, Charron DF, Role of migratory birds in introduction and range expansion of Ixodes scapularis ticks and of Borrelia burgdorferi and Anaplasma phagocytophilum in Canada. Appl Environ Microbiol. 2008;74:1780–90. DOIPubMedGoogle Scholar
- Bouchard C, Leighton PA, Beauchamp G, Nguon S, Trudel L, Milord LF, Harvested white-tailed deer as sentinel hosts for early establishing Ixodes scapularis populations and risk from vector-borne zoonoses in southeastern Canada. J Med Entomol. 2013;50:384–93. DOIPubMedGoogle Scholar
- Dibernardo A, Cote T, Ogden NH, Lindsay RL. The prevalence of Borrelia miyamotoi infection, and co-infections with other Borrelia spp. in Ixodes scapularis ticks collected in Canada. Parasites & Vectors. 2014 7:183.
- Massung RF, Courtney JW, Hiratzka SL, Pitzer VE, Smith G, Dryden RL. Anaplasma phagocytophilum in white-tailed deer. Emerg Infect Dis. 2005;11:1604–6. DOIPubMedGoogle Scholar
- Kim E-J, Bauer C, Grevelding CG, Quack T. Improved PCR/nested PCR approaches with increased sensitivity and specificity for the detection of pathogens in hard ticks. Ticks Tick Borne Dis. 2013;4:409–16.
- Campsall PA, Au NHC, Prendiville JS, Speert DP, Tan R, Thomas EE. Detection and genotyping of varicella-zoster virus by Taqman allelic discrimination real-time PCR. 2004;42:109–13.
Figures
Tables
Cite This ArticleTable of Contents – Volume 20, Number 12—December 2014
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Neil Chilton, Department of Biology, University of Saskatchewan, 112 Science Pl, Saskatoon, SK. S7N 5E2, Canada
Top