Hepacivirus Infection in Domestic Horses, Brazil, 2011–2013
Bernard Salame Gemaque, Alex Junior Souza de Souza, Manoel do Carmo Pereira Soares, Andreza Pinheiro Malheiros, Andrea Lima Silva, Max Moreira Alves, Michele Soares Gomes-Gouvêa, João Renato Rebello Pinho, Heriberto Ferreira de Figueiredo, Djacy Barbosa Ribeiro, Jonan Souza da Silva, Leopoldo Augusto Moraes, Ana Silvia Sardinha Ribeiro, and Washington Luiz Assunção Pereira
Author affiliations: Universidade Federal Rural da Amazônia, Belém, Brazil (B.S. Gemaque, H.F. de Figueiredo, D.B. Ribeiro, J.S. da Silva, A.S.S. Ribeiro, W.L.A. Pereira); Instituto Evandro Chagas, Belém (B.S. Gemaque, A.J.S. de Souza, M.C.P. Soares, A.P. Malheiros, A.L. Silva, M.M. Alves); Universidade de São Paulo, São Paulo, Brazil (A.J.S. de Souza, M.S. Gomes-Gouvêa, J.R.R. Pinho); Universidade Federal do Pará, Belém (L.A. Moraes)
Main Article
Figure
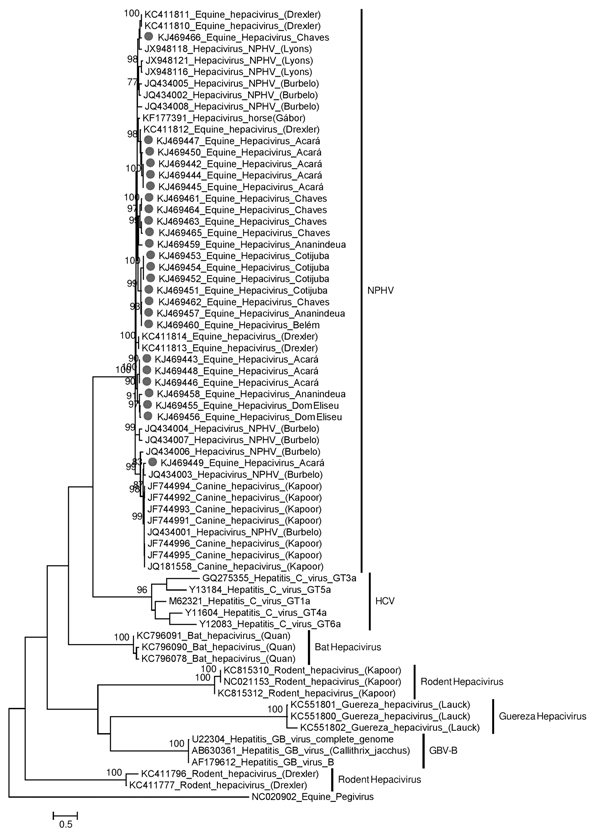
Figure. Maximum-likelihood phylogenetic tree of the partial nucleotide sequences of the nonstructural protein 3 region (294 bp) of Hepacivirus. The retrieved sequences from GenBank are indicated by the accession number followed by the species from which each was isolated. The 25 sequences obtained from 300 equids in 7 cities and 1 district of the State of Pará, Brazil, during January 2011–November 2013, are indicated with a dot and are identified by their GenBank accession numbers followed by their community of origin. Bootstrap values (1,000 replicates) >70% are listed at the nodes. One sequence of equine Pegivirus was used as an outgroup. GBV-B, GB virus B; HCV, hepatitis C virus; NPHV, nonprimate hepaciviruses. Scale bar indicates nucleotide substitutions per site.
Main Article
Page created: November 05, 2014
Page updated: November 05, 2014
Page reviewed: November 05, 2014
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
