Volume 20, Number 12—December 2014
Letter
Third Strain of Porcine Epidemic Diarrhea Virus, United States
Figure
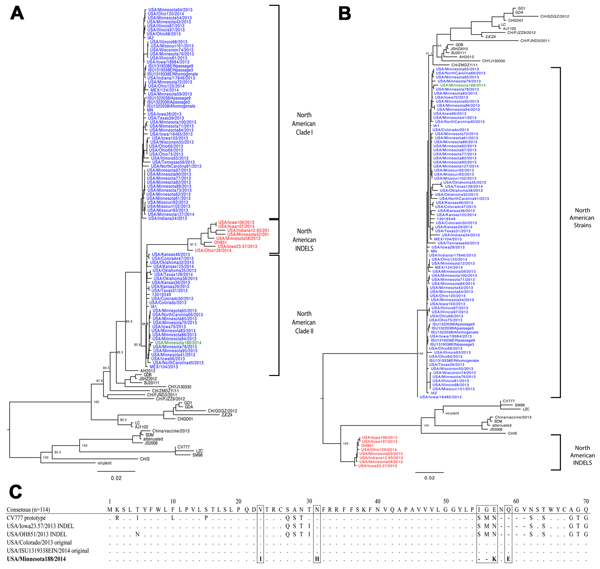
Figure. Complete genome (A) and spike gene (B) phylogenetic analysis using the maximum likelihood method with the general time reversible (GTR) nucleotide substitution model. Green indicates Minnesota188 strain, red indicates North American INDEL strains, and blue indicates the remaining North American porcine epidemic diarrhea virus (PEDV) strains. Scale bar indicates percentage of dissimilarity between sequences. C) Spike gene alignment of first 70 aa). The consensus represents 114 sequences used in the analysis. PEDV prototype strain CV777, 2 American original and INDEL PEDV strains are illustrated. Dots indicate amino acid residues matching the consensus are represented by dots; dashes represent amino acid deletions.
Page created: November 19, 2014
Page updated: November 19, 2014
Page reviewed: November 19, 2014
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.