Volume 20, Number 3—March 2014
Research
Monitoring Water Sources for Environmental Reservoirs of Toxigenic Vibrio cholerae O1, Haiti
Abstract
An epidemic of cholera infections was documented in Haiti for the first time in more than 100 years during October 2010. Cases have continued to occur, raising the question of whether the microorganism has established environmental reservoirs in Haiti. We monitored 14 environmental sites near the towns of Gressier and Leogane during April 2012–March 2013. Toxigenic Vibrio cholerae O1 El Tor biotype strains were isolated from 3 (1.7%) of 179 water samples; nontoxigenic O1 V. cholerae was isolated from an additional 3 samples. All samples containing V. cholerae O1 also contained non-O1 V. cholerae. V. cholerae O1 was isolated only when water temperatures were ≥31°C. Our data substantiate the presence of toxigenic V. cholerae O1 in the aquatic environment in Haiti. These isolations may reflect establishment of long-term environmental reservoirs in Haiti, which may complicate eradication of cholera from this coastal country.
Epidemic cholera was identified during October 2010 in Haiti; initial cases were concentrated along the Artibonite River (1,2). The clonal nature of isolates during this initial period of the epidemic has been described (3–6). Because cholera had not been reported in Haiti for at least 100 years, there is a high likelihood that the responsible toxigenic Vibrio cholerae strain was introduced into Haiti, possibly through Nepalese peacekeeping troops garrisoned at a camp along the Artibonite River (4,7). In the months after October 2010, cholera spread quickly through the rest of Haiti: 604,634 cases and 7,436 deaths were reported in the first year of the epidemic (1). In the intervening years, cases and epidemics have been reported, and it has been suggested that onset of the rainy season serves as a trigger for disease occurrences (2,8).
V. cholerae is well recognized as an autochthonous aquatic microorganism species with the ability to survive indefinitely in aquatic reservoirs and is possibly in a “persister” phenotype (9). V. cholerae strains can also persist in aquatic reservoirs as a rugose variant that promotes formation of a biofilm that confers resistance to chlorine and to oxidative and osmotic stresses (10–13) and also persists in a viable but nonculturable form (14). Work by our group and others suggests that cholera epidemics among humans are preceded by an environmental bloom of the microorganism and subsequent spillover into human populations (15–17). In our studies in Peru (16), water temperature was found to be the primary trigger for these environmental blooms and could be correlated with subsequent increases in environmental counts and occurrence of human illness.
To understand patterns of ongoing cholera transmission and seasonality of cholera in Haiti, and to assess the likelihood of future epidemics, it is essential to know whether environmental reservoirs of toxigenic V. cholerae O1 have been established, where these reservoirs are located, and what factors affect the occurrence and growth of the microorganism in the environment. We report the results of an initial year of monitoring of environmental sites in the Ouest Department of Haiti, near the towns of Leogane and Gressier, where the University of Florida (Gainesville, FL, USA) has established a research laboratory and field area.
Environmental Sampling Sites
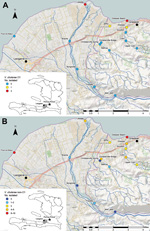
Fifteen fixed environmental sampling sites were selected near Gressier and Leogane (Figures 1,2). Sites were selected along transects of 3 rivers in the area and at 1 independent estuarine site: the Momance River (4 up-river sites and 1 estuarine site at the mouth of the river), the Gressier River (4 up-river sites and1 estuarine site at the mouth of the river), the Tapion River (4 river sites), and an independent estuarine site at Four-a-chaux, which is a historic ruin and tourist attraction. All sites were >0.5 miles apart, with the exception of the Christianville Bridge and Spring sites, which were 0.25 miles apart. Topography of this area is typical for Haiti: rivers originated in the mountains (peaks in the region are >8,000 feet) and flowed into a broad flood plain where Gressier and Leogane were located. Up-river sites on the Momance and Gressier Rivers were in the mountains, where human populations are limited. Water samples were collected once a month from each site during April 2012–March 2013. A total of 179 samples were collected for culture for V. cholerae; 176 samples were available for measurement of water quality parameters.
Isolation and Identification of V. cholerae from Environmental Sites
For the isolation of V. cholerae, 500 ml of water was collected in a sterile 500-mL Nalgene (http://nalgene.com/) bottle from each fixed site; the samples were transported at ambient temperature to the University of Florida laboratory at Gressier and processed for detection of V. cholerae within 3 hours of collection.
In addition to the conventional sample enrichment technique (18), we used alkaline peptone water (APW) to enrich water samples. A 1.5-mL water sample was enriched with 1.5 mL of 2× APW in 3 tubes: 1 tube was incubated at 37°C for 6–8 hours (18), another tube was incubated overnight at 37°C, and the third tube was incubated at 40°C for 6–8 hours. Subsequently, a loopful of culture from each tube was streaked onto thiosulfate citrate bile salts sucrose agar (Becton-Dickinson, Franklin Lakes, NJ, USA), and the plates were incubated overnight at 37°C. From each plate, 6–8 yellow colonies exhibiting diverse morphology were transferred to L-agar; these plates were incubated overnight at 37°C. Each colony was examined by using the oxidase test; oxidase-positive colonies were tested by using V. cholerae O1–specific polyvalent antiserum and O139-specific antiserum (DENKA SEIKEN Co., Ltd, Tokyo, Japan). The isolates were further examined by using colony PCR for the presence of ompW and toxR genes specific for V. cholerae spp. as described (9).
Screening of Aquatic Animals and Plants
To determine whether they serve as reservoirs for V. cholerae O1, we collected aquatic animals typically eaten by humans, including shrimp, fish, crab, crayfish, and aquatic plants (n = 144) weekly during February 5–22, 2013. The samples were collected from 14 environmental sites. Each sample was placed into a sterile plastic seam–locking bag and transported to the laboratory. One gram of the sample was mixed with 100 mL of saline and then homogenized in a sterile blender; 1.5 ml of the resultant mixture was enriched in 2× APW and processed as described.
Genetic Characterization of V. cholerae O1 Strains
To further characterize the environmental V. cholerae O1 serogroup Ogawa biotype El Tor strains, we subjected all V. cholerae O1 isolates from water and seafood to PCR analysis for key virulence genes, including ctxA, ctxB-CL, (MAMA-CL), ctxB-ET (MAMA-ET), rstR-ET, rstR-CL, rstC-ET, rstC-CL, tcpA-CL, and tcpA-ET, as described (19,20). The chromosomal DNA was extracted from each strain by using a GenElute Bacterial Genomic DNA kit (Sigma-Aldrich, St. Louis, MO, USA), and the DNA was used for PCR templates; the PCR conditions were as described (3).
Aerobic Plate Counts
To determine total aerobic bacterial counts in water samples, we plated undiluted, 10- and 100-fold dilutions of water onto L-agar and incubated overnight at 37°C. The countable plates (100–300 colonies) were used to determine the total (CFU/mL) culturable bacteria present in the water samples.
Water Parameters, Rainfall, and Human Case Counts
When collecting water samples, we measured physical parameters, including pH, water temperature, dissolved oxygen, total dissolved solids, salinity, and conductivity in the field sites by using a HACH portable meter (HACH Company, Loveland, CO, USA) and designated electrodes following the manufacturer’s recommendations. Rainfall estimates were based on National Aeronautics and Space Administration data for the study region bounded by the rectangle (18.2°–18.6°N, 17.1°–17.8°W) by using the average daily rainfall measurement tool, Tropical Rainfall Measuring Mission 3B42_daily (21). Estimates of average precipitation (mm/day) with a spatial resolution of 0.25 × 0.25 degrees were aggregated to obtain weekly accumulated rainfall measurements during the study period. Cholera incidence data were obtained from daily reports by Ouest Department (excluding Port-au-Prince) to the Haitian Ministry of Public Health and Population and aggregated to total cases per week during April 20, 2012–March 27, 2013 (22).
Data Analysis
We examined the effects of water quality factors on the presence of toxigenic and nontoxigenic V. cholerae by conditional logistic regression after stratification for the site. Stratification excluded sites that had all-positive or all-negative outcomes; of the remaining sites, regression analysis showed O1 V. cholerae in 47 observations from 4 sites and non-O1/non-O139 V. cholerae in 154 observations from13 sites. As shown in Figure 1, we performed cartography by using ArcGIS version 10 (ESRI, Redlands, CA, USA).
V. cholerae O1 serogroup Ogawa biotype El Tor was isolated from 6 (3.4%) of the 179 water samples and 1 (0.7%) of the 144 aquatic animal and plant samples by using modified APW enrichment techniques. Of those 7 environmental isolates, 3 (43%) were confirmed as ctx-positve toxigenic V. cholerae O1 strains, and 4 (57%) were confirmed as ctx-negative V. cholerae O1 strains by using genetic analysis as described below. As shown in Table 1, APW enrichment at 37°C overnight or incubation at 40°C for 6–8 hours, or both, enhanced the rate of isolation of V. cholerae O1 from samples. PCR analysis of the key virulence genes showed that 3 (43%) of the 7 isolates, all from water, were positive for key virulence genes, including cholera toxin genes and tcpA genes, and that 4 (57%) isolates exhibited no cholera toxin bacteriophage (CTXΦ)–related genes (23; Table 2). To further assess the PCR results, we sequenced DNA flanking the CTXΦ from 1 strain, Env-9 (Table 2). Sequence data corroborated PCR results that indicated that Env-9 lacked CTXΦ.
Physical parameters for the environmental water samples are summarized in Table 3. Because the sites varied from mountains to floodplain to estuaries, there was relatively wide variability in salinity (0–21.6 g/L), pH (6.4–8.6), and temperature (24.3–33.7°C). Temperatures tended to increase as rivers approached the sea. As shown in Figure 2, mean water temperature from all sites showed evidence of seasonal variation. Measurement of rainfall was available for the region as a whole (Figure 3). However, site-specific rainfall data were not available; consequently, rainfall was not included in the regression models.
Isolation of V. cholerae O1 strains was most common from the sites at the mouths of the Momance and Gressier Rivers (Figure 1, panel A). In a conditional logistic regression analysis with water quality factors (Table 4), the only variable that emerged as statistically significant was water temperature (odds ratio 2.14, 95% CI 1.06–4.31); all isolations of V. cholerae O1 (toxigenic and nontoxigenic) occurred at water temperatures of >31°C. As shown in Figure 3, there was evidence that V. cholerae O1 isolation was more common in the environment preceding epidemic peaks of disease among humans; however, numbers of isolations were too small to permit statistical analysis. Of 179 samples, the only V. cholerae O1 isolate from aquatic animals or plants was from a shrimp sample and was nontoxigenic; it was collected simultaneously with a water sample that was also positive for nontoxigenic V. cholerae O1.
Non-O1 V. cholerae was much more common in the environment than V. cholerae O1 strains and was isolated from 56 (31%) of 179 water samples. As observed with O1 strains, isolations were more common at the mouths of the rivers and in estuarine areas (Figure 1, panel B); however, the non-O1 strain was found farther upriver than were O1 strains and was isolated from several sites in the mountains. Non-O1 strains were isolated from all sites that were also positive for O1 strains. Non-O1 strains were isolated in all months, without an obvious association with regional rainfall totals or cholera incidence. In a conditional logistic regression analysis, isolation of non-O1 strains was significantly associated (p<0.05) with higher water temperature and moderate levels of dissolved oxygen (Table 4).
Before this study, isolation of 2 toxigenic V. cholerae O1 strains from large-volume water samples (30 L) was reported in the Artibonite region (24); other studies at that time suggested that V. cholerae O1 strains were not present, or present at only minimal levels (2,25) in the environment in Haiti. In contrast, we isolated ctx-positive and ctx-negative V. cholerae O1 serogroup Ogawa biotype El Tor strains (Table 1) in the environment at a frequency comparable to that reported from cholera-endemic areas such as Bangladesh (17). Our successful isolation of the microorganism from the environment may reflect localization of environmental isolates near Gressier and Leogane, where our study was conducted; however, we believe that our findings are more likely to be a reflection of the method used. Data presented here suggest that, in addition to conventional APW enrichment, longer APW enrichment time and enrichment at higher temperatures contributed to an increased rate of isolation of V. cholerae O1 strains from aquatic environmental reservoirs (Table 1), resulting in successful isolation from 1.5-mL water samples. We also note some issues relating to sample transport: Baron et al. (25) transported their water samples on ice in coolers; our samples were transported at room temperature. As has been reported, Vibrio spp. are extremely sensitive to low temperatures (26), and in our experience, transport of samples on ice resulted in a marked reduction in isolation rates.
Water from which we isolated V. cholerae spp. tended to have been sampled at the point where rivers meet the sea, and in adjacent estuarine areas, again following the patterns reported from Bangladesh (17). Water temperature was found to be the single physical parameter that was substantially associated with isolation of these organisms; higher temperatures were concentrated downriver and in estuarine areas. For our analysis, we used a conditional logistic regression model to permit stratification by site. Although we found very low numbers for V. cholerae O1 isolates (6 positive water samples), results coincided with the non-O1 results and the exploratory data analysis. In our studies of aquatic animals likely to be eaten by humans, we did isolate V. cholerae O1 from shrimp in 1 instance. The isolate was nontoxigenic; consequently, its association with disease is unclear.
After analyzing the results of this study, we asked the following question: has V. cholerae O1 become established in environmental reservoirs in Haiti? Toxigenic V. cholerae O1 strains are clearly present in the environment, and it may be that the isolates that we identified are the result of fecal contamination of the environment by persons infected with V. cholera strains. Although data are limited, there was at least a suggestion that isolation of V. cholera strains from environmental reservoirs was more common at the beginning of epidemic spikes of human disease (as has been described in association with environmental reservoirs) (16,17,27) rather than at the height of epidemics among humans, as might have been expected related to fecal contamination. We also found non-O1 strains widely distributed throughout the environment, including mountain river sites, consistent with widespread dissemination in environmental reservoirs. Although we cannot be certain that O1 and non-O1 strains grow under comparable conditions, the clear establishment of non-O1 V. cholerae strains in environmental reservoirs suggests that conditions are appropriate for growth of V. cholerae O1 strains. Of potentially greater interest is the observation that only 3 of the 7 (47%) V. cholerae O1 biotype El Tor strains isolated carried the ctx genes (Table 2). Data from 1 ctx-negative strain (Env-9) was consistent with absence of the entire CTXΦ. We propose that the 3 isolates that are positive for ctx genes be classified as circulating V. cholerae altered biotype El Tor strains in Haiti. To better understand the evolutionary mechanisms involved, we are performing further sequence analysis of clinical and environmental strains.
The apparent introduction of toxigenic V. cholerae O1 in Haiti in 2010, after decades during which no cholera cases were reported, was unquestionably a public health disaster. If these O1 strains establish stable environmental reservoirs in Haiti, in the setting of ongoing problems with water and sanitation, there is a high likelihood that we will see recurrent epidemics within the country. These circumstances clearly have implications for current plans by the Haitian Ministry of Public Health to eradicate cholera in Haiti within a decade (28). The proposed implementation of vaccination programs and efforts to improve water supplies and sanitation will undoubtedly reduce case numbers, but as long as the causative microorganism is present in the environment, eradication of the disease will not be possible. Establishment of environmental reservoirs and recurrent epidemics may also serve as a potential source for transmission of the disease to the Dominican Republic and other parts of the Caribbean (1). Ongoing monitoring of potential environmental reservoirs in the areas near Gressier and Leogane as well as in sentinel sites throughout the country will be necessary to assess this risk and to permit development of rational public health interventions for cholera control.
Mr Alam is a research scholar at the Department of Environmental and Global Health in the College of Public Health and Health Professions, University of Florida at Gainesville. His research interests focus on the ecology and epidemiology of V. cholerae.
Acknowledgments
We thank Mohammad Jubair for his technical help with this study.
This work was supported in part by National Institutes of Health grants RO1 AI097405 awarded to J.G.M. and a Department of Defense grant (C0654_12_UN) awarded to A.A.
References
- Barzilay EJ, Schaad N, Magloire R, Mung KS, Boncy J, Dahourou GA, Cholera surveillance during the Haiti epidemic — the first 2 years. N Engl J Med. 2013;368:599–609 and. DOIPubMedGoogle Scholar
- Gaudart J, Rebaudet S, Barrais R, Boncy J, Faucher B, Piarroux M, Spatio-temporal dynamics of cholera during the first year of the epidemic in Haiti. PLoS Negl Trop Dis. 2013;7:e2145 . DOIPubMedGoogle Scholar
- Ali A, Chen Y, Johnson JA, Redden E, Mayette Y, Rashid MH, Recent clonal origin of cholera in Haiti. Emerg Infect Dis. 2011;17:699–701 and. DOIPubMedGoogle Scholar
- Chin C-S, Sorenson J, Harris JB, Robins WP, Charles RC, Jean-Charles RR, The origin of the Haitian cholera outbreak strain. N Engl J Med. 2011;364:33–42 and. DOIPubMedGoogle Scholar
- Talkington D, Bopp C, Tarr C, Parsons MB, Dahourou G, Freeman M, Characterization of toxigenic Vibrio cholerae from Haiti, 2010–2011. Emerg Infect Dis. 2011;17:2122–9 .PubMedGoogle Scholar
- Katz LS, Petkau A, Beaulaurier J, Tyler S, Antonova ES, Turnsek MA, Evolutionary dynamics of Vibrio cholerae O1 following a single-source introduction to Haiti. MBio. 2013;4:00398–00413 . DOIPubMedGoogle Scholar
- Enserink M. Haiti's cholera outbreak. Cholera linked to U.N. forces, but questions remain. Science. 2011;332:776–7 and . DOIPubMedGoogle Scholar
- Rinaldo A, Bertuzzo E, Mari L, Righetto L, Blokesch M, Gatto M, Reassessment of the 2010–2011 Haiti cholera outbreak and rainfall-driven multiseason projections. Proc Natl Acad Sci U S A. 2012;109:6602–7 and. DOIPubMedGoogle Scholar
- Jubair M, Morris JG, Ali A. Survival of Vibrio cholerae in nutrient-poor environments is associated with a novel “persister” phenotype. PLoS ONE. 2012;7:e45187 and. DOIPubMedGoogle Scholar
- Rice EW, Johnson CJ, Clark RM, Fox KR, Reasoner DJ, Dunnigan ME, Chlorine and survival of “rugose” Vibrio cholerae. Lancet. 1992;340:740 and. DOIPubMedGoogle Scholar
- Ali A, Rashid MH, Karaolis DKR. High-frequency rugose exopolysaccharide production by Vibrio cholerae. Appl Environ Microbiol. 2002;68:5773–8 and. DOIPubMedGoogle Scholar
- Yildiz FH, Schoolnik GK. Vibrio cholerae O1 El Tor: identification of a gene cluster required for the rugose colony type, exopolysaccharide production, chlorine resistance, and biofilm formation. Proc Natl Acad Sci U S A. 1999;96:4028–33 and. DOIPubMedGoogle Scholar
- Wai SN, Mizunoe Y, Takade A, Kawabata SI, Yoshida SI. Vibrio cholerae O1 strain TSI-4 produces the exopolysaccharide materials that determine colony morphology, stress resistance, and biofilm formation. Appl Environ Microbiol. 1998;64:3648–55 .PubMedGoogle Scholar
- Colwell RR, Huq A. Vibrios in the environment: viable but nonculturable Vibrio cholerae. In: Wachsmuth IK, Blake PA, Olsvik Ø, editors. Vibrio cholerae and cholera: molecular to global perspectives. Washington (DC): American Society for Microbiology; 1994.
- Morris JG Jr. Cholera–modern pandemic disease of ancient lineage. Emerg Infect Dis. 2011;17:2099–104 and. DOIPubMedGoogle Scholar
- Franco AA, Fix AD, Prada A, Paredes E, Palomino JC, Wright AC, Cholera in Lima, Peru, correlates with prior isolation of Vibrio cholerae from the environment. Am J Epidemiol. 1997;146:1067–75 and. DOIPubMedGoogle Scholar
- Huq A, Sack RB, Nizam A, Longini IM, Nair GB, Ali A, Critical factors influencing the occurrence of Vibrio cholerae in the environment of Bangladesh. Appl Environ Microbiol. 2005;71:4645–54 and. DOIPubMedGoogle Scholar
- Lesmana M, Rockhill RC, Sutanti D, Sutomo A. An evaluation of alkaline peptone water for enrichment of Vibrio cholerae in feces. Southeast J. Trop. Med. Public Health. 1985;16:265–7.PubMedGoogle Scholar
- Morita M, Ohnishi M, Arakawa E, Bhuiyan NA, Nusrin S, Alam M, Development and validation of a mismatch amplification mutation PCR assay to monitor the dissemination of an emerging variant of Vibrio cholerae O1 biotype El Tor. Microbiol Immunol. 2008;52:314–7 and. DOIPubMedGoogle Scholar
- Aliabad NH, Bakhshi B, Pourshafie MR, Sharifnia A, Ghorbani M. Molecular diversity of CTX prophage in Vibrio cholerae. Lett Appl Microbiol. 2012;55:27–32 and. DOIPubMedGoogle Scholar
- National Aeronautics and Space Administration. NASA earth data. Goddard Earth Sciences Data and Information Services Center. (search keyword: 3B42 V7 derived). 2013 [cited 2013 May 14]. http://mirador.gsfc.nasa.gov/
- Haitian Ministry of Public Health and Population. Daily reports of cholera cases by commune. May 2013 [in French] [cited 2013 May 14]. http://mspp.gov.ht/newsite/documentation.php
- Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–4 and. DOIPubMedGoogle Scholar
- Hill VR, Cohen N, Kahler AM, Jones JL, Bopp CA, Marano N, Toxigenic Vibrio cholerae O1 in water and seafood, Haiti. Emerg Infect Dis. 2011;17:2147–50 and. DOIPubMedGoogle Scholar
- Baron S, Lesne J, Moore S, Rossignol E, Rebaudet S, Gazin P, No evidence of significant levels of toxigenic V. cholerae O1 in the Haitian aquatic environment during the 2012 rainy season. PLoS Curr. 2013;13:1–14 .PubMedGoogle Scholar
- Huq A, West PA, Small EB, Huq MI, Colwell RR. Influence of water temperature, salinity, and pH on survival and growth of toxigenic Vibrio cholerae serovar O1 associated with live copepods in laboratory microcosms. Appl Environ Microbiol. 1984;48:420–4 .PubMedGoogle Scholar
- Dalsgaard A, Serichantalergs O, Forslund A, Lin W, Mekalanos J, Mintz E, Clinical and environmental isolates of Vibrio cholerae serogroup O141 carry the CTX phage and the genes encoding the toxin-coregulated pili. J Clin Microbiol. 2001;39:4086–92 and. DOIPubMedGoogle Scholar
- Haitian Ministry of Public Health and Population. National plan for the elimination of cholera in Haiti 2013–2022. 2012 Feb [cited 2013 May 14]. http://www.paho.org/hq/index.php?option=com_docman&task=doc_view&gid=20326&Itemid=270&lang=en
Figures
Tables
Cite This ArticleTable of Contents – Volume 20, Number 3—March 2014
| EID Search Options |
|---|
|
|
|
|
|
|


Please use the form below to submit correspondence to the authors or contact them at the following address:
Afsar Ali, Department of Environmental and Global Health, School of Public Health and Health Professions, Emerging Pathogens Institute, University of Florida at Gainesville, 2055 Mowry Rd, Gainesville, FL 32610, USAAfsar Ali, Department of Environmental and Global Health, School of Public Health and Health Professions, Emerging Pathogens Institute, University of Florida at Gainesville, 2055 Mowry Rd, Gainesville, FL 32610, USA
Top