Volume 20, Number 5—May 2014
Dispatch
Full-Genome Analysis of Avian Influenza A(H5N1) Virus from a Human, North America, 2013
Figure 1
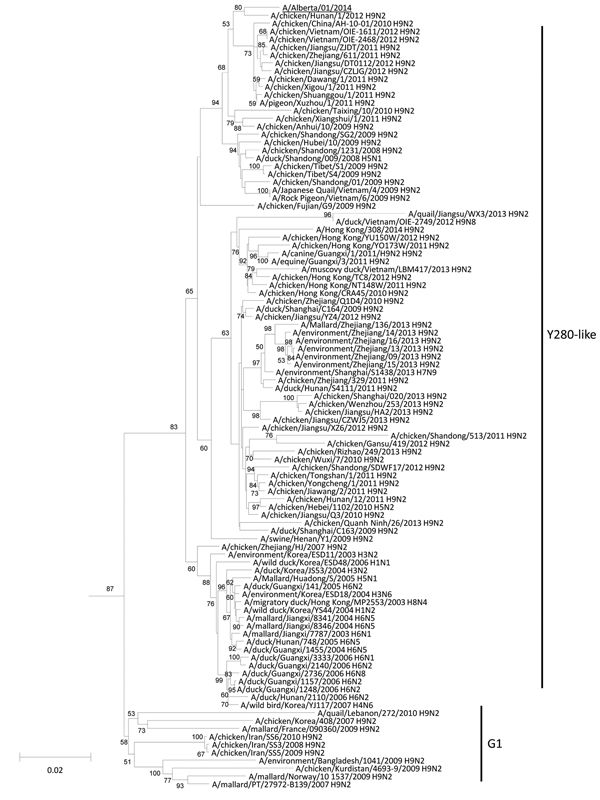
Figure 1. Neighbor-joining phylogenetic tree of the polymerase basic 2 (PB2) genes of H9N2 subtype lineage avian influenza A viruses with A/Alberta/01/2014 (GISAID accession noEPI500778)The avian influenza A(H5N1) virus detected in Canada is underlinedMajor lineages of the H9N2 subtype–like PB2 genes are depicted to the right of the phylogenetic clustersBootstraps generated from 1,000 replicates are shown at branch nodesScale bar represents nucleotide substitutions per siteGSAID, Global Initiative on Sharing Avian Influenza Data.
Page created: April 17, 2014
Page updated: April 17, 2014
Page reviewed: April 17, 2014
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.