Volume 20, Number 6—June 2014
Research
Bats as Reservoir Hosts of Human Bacterial Pathogen, Bartonella mayotimonensis
Figure 2
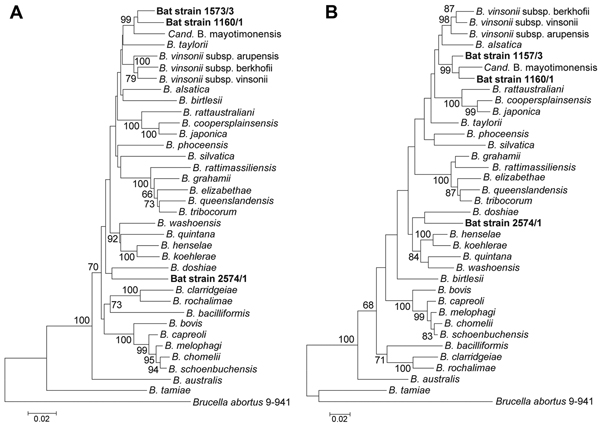
Figure 2. Phylogenetic positions of the bat blood isolates among members of the genus BartonellaNeighbor-joining (A) and maximum-likelihood (B) trees are based on the alignment of concatenated sequences of 4 multilocus sequence analysis markers (rpoB, gltA, 16S rRNA, and ftsZ)Sequence information from the type strains of all known Bartonella species and from the Candidatus Bmayotimonensis human strain was included into the analysis (Technical Appendix Table 5)Numbers on branches indicate bootstrap support values derived from 1,000 tree replicasBootstrap values >60 are shownScale bar indicates nucleotide substitutions per site.
1These authors contributed equally to this article.
Page created: May 16, 2014
Page updated: May 16, 2014
Page reviewed: May 16, 2014
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.