Volume 20, Number 6—June 2014
Dispatch
New Hepatitis E Virus Genotype in Camels, the Middle East
Figure 2
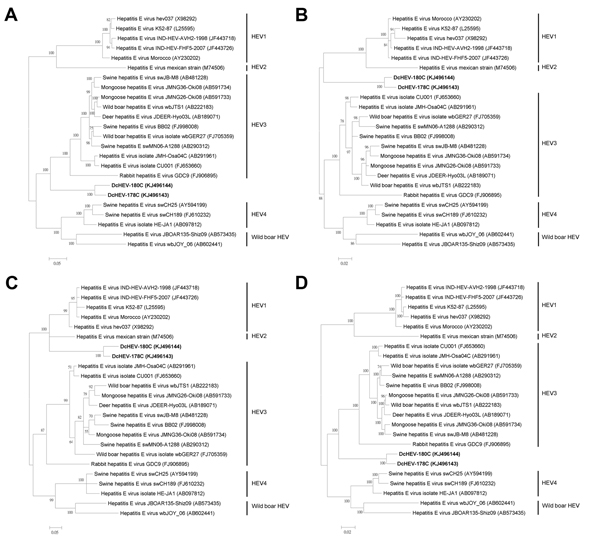
Figure 2. Phylogenetic analyses of open reading frame (ORF) 1 (A), ORF2 (B), ORF3 (C), and ORF1/ORF2 proteins, excluding the hypervariable region (HVR) (D) of hepatitis E virus (HEV) from dromedary camels (DcHEV)The trees were constructed by using Bayesian methods of phylogenetic reconstruction (www.fifthdimension.jp/products/mrbayes5d/), and ProtTest-suggested JTT+I+G+F, MtMam+I+G+F, HIVw+I+G+F, and JTT+I+G+F (http://darwin.uvigo.es/software/prottest.html) are the optimal substitution models for ORF1, ORF2, ORF3, and concatenated ORF1/ORF2 excluding HVR, respectivelyFor this analysis we included amino acid positions 1698, 660, 113, and 2282 in ORF1, ORF2, ORF3 and concatenated ORF1/ORF2 excluding HVR, respectivelyFor ORF2 and concatenated ORF1/ORF2 excluding HVR, the scale bars indicate the estimated number of substitutions per 50 aaFor ORF1 and ORF3, the scale bars indicate the estimated number of substitutions per 20 aaBoldface indicates the 2 strains of DcHEV with complete genomes sequenced in this study.
1These authors contributed equally to this article.