Volume 21, Number 1—January 2015
Research
Clinical, Environmental, and Serologic Surveillance Studies of Melioidosis in Gabon, 2012–2013
Abstract
Burkholderia pseudomallei, an environmental gram-negative bacillus, is the causative agent of melioidosis and a bio-threat agent. Reports of B. pseudomallei isolation from soil and animals in East and West Africa suggest that melioidosis might be more widely distributed than previously thought. Because it has been found in equatorial areas with tropical climates, we hypothesized that B. pseudomallei could exist in Gabon. During 2012–2013, we conducted a seroprevalance study in which we set up microbiology facilities at a large clinical referral center and prospectively screened all febrile patients by conducting blood cultures and testing for B. pseudomallei and related species; we also determined whether B. pseudomallei could be isolated from soil. We discovered a novel B. pseudomallei sequence type that caused lethal septic shock and identified B. pseudomallei and B. thailandensis in the environment. Our data suggest that melioidosis is emerging in Central Africa but is unrecognized because of the lack of diagnostic microbiology facilities.
The Tier 1 bio-threat agent Burkholderia pseudomallei is an environmental gram-negative bacillus and the cause of melioidosis, a disease characterized by sepsis, pneumonia, and abscess formation in almost any organ (1–3). B. thailandensis is closely related to B. pseudomallei but rarely causes disease in humans or animals; it is usually distinguished from B. pseudomallei by its ability to assimilate arabinose (4–6). Melioidosis mainly affects those who are in regular contact with soil and water and is associated with a mortality rate of up to 40% in resource-poor environments. The major regions to which melioidosis is endemic are Southeast Asia and tropical Australia (1,2). The northern tip of the Northern Territory in Australia and northeast Thailand represent hot spots, where annual incidence is up to 50 cases per 100,000 persons (1,7).
The emergence of melioidosis in Brazil is an example of increasing recognition of the disease in areas where it is probably endemic, and cases have become apparent as a result of enhanced awareness and diagnostics (1,8). Human B. pseudomallei infection has been reported from Malawi, Nigeria, The Gambia, Kenya, and Uganda; however, human cases in Africa seem to be few and isolated, although this finding could be the result of underrecognition and underreporting (1,9–12). Although reports of B. pseudomallei isolation from soil and animals in East and West Africa are limited, they suggest that melioidosis could be widely distributed across this region (13,14).
Given the equatorial tropical distribution of B. pseudomallei and B. thailandensis, we hypothesized that these bacteria are present in the central African country of Gabon, potentially causing disease. By conducting a seroprevalence study, an environmental survey, and setting up microbiology facilities for B. pseudomallei detection at a large referral hospital, we detected B. pseudomallei in soil and identified it as a cause of lethal infection in Gabon. We also detected B. thailandensis in environmental soil samples, indicating that this organism is also present in Gabon.
Study Sites and Populations
The study was performed in Moyen-Ogooué and Ngounié Provinces (combined population 162,000) in central Gabon; these 2 provinces cover an area of 56,285 km2 and consist of predominantly dense primary rain forest. For the seroprevalence surveillance study, 304 serum samples were collected from healthy nonfebrile school children (12–20 years of age) living in and around Lambaréné, the capital of Moyen-Ogooué Province; these children also participated in a chemoprophylaxis study for malaria (15).
A prospective analysis of community-acquired bloodstream infections was performed at Albert Schweitzer Hospital (which admits ≈6,000 patients annually) in Lambaréné (population 24,000), in the Central African rain forest on the river Ogooué, Gabon. The rainy season starts in October and ends in June (including a short dry season in December–January). Mean annual rainfall is 1,981 mm (78 inches), which is equivalent to that in northeastern Thailand (16). Studies were approved by the Centre National de la Recherche Scientifique et Technologique, Libreville, and the scientific review committee of the Centre de Recherches Médicales de Lambaréné, Albert Schweitzer Hospital.
Prospective Analysis of Community-Acquired Bloodstream Infections
To obtain data about the prevalence and causes of community-acquired bloodstream infections in Lambaréné, we prospectively monitored all blood cultures for febrile patients admitted to Albert Schweitzer Hospital for 1 year (June 1, 2012–May 31, 2013) by using BacT/Alert PF (bioMérieux, Marcy l'Etoile, France). Criteria for ordering blood cultures were left to the discretion of the treating physician. Technicians and staff of the clinical microbiology laboratory received additional training on sample handling and processing (17,18). All oxidase-positive, gram-negative bacteria that were not Pseudomonas aeruginosa were further tested to determine whether they were B. pseudomallei by using the subculture and identification methods described below. Antimicrobial drug susceptibilities were determined by using Etest (bioMérieux) on Mueller-Hinton-agar (bioMérieux); when available, break points were defined as described (19).
B. pseudomallei Antibody Detection by Indirect Hemagglutination Assay
During May 2012, presence and titer of antibodies to B. pseudomallei in healthy schoolchildren were determined using by the indirect hemagglutination assay (IHA) as described (20,21), with pooled antigens prepared from 2 B. pseudomallei isolates from Thailand. An antibody titer of ≥1:40 was used as the cutoff value for seropositivity (22).
Soil Sampling Study
During July 2012–September 2012, soil sampling to test for the presence of B. pseudomallei was based on consensus guidelines, and direct culture of soil in enrichment broth was performed (17,23). A total of 8 sites around the residences of children were selected on the basis of local maps and consultations with inhabitants throughout the provinces of Moyen-Ogooué (6 sites) and Ngounié (2 sites) and on known factors associated with the presence of B. pseudomallei (e.g., wet soil such as rice paddies or land use such as goat farming) (17) (Figure 1). Within each sampling area (50 × 50 m2), a fixed-interval sampling grid was used to collect 100 samples per field, 5 m apart. For each sample, 10 g of soil was collected from a depth of 30 cm, stored away from direct sunlight, and processed within 3 h.
Isolation of potential Burkholderia spp. from soil was performed as described (17,23). In brief, 10 g of soil was diluted in 10 mL of threonine–basal salt solution plus colistin at 50 mg/liter (TBSS-C50 broth) containing crystal violet and was vortexed for 30 s before incubation at ≈42°C for 48 h. Ten μL of supernatant was subcultured onto Ashdown-agar and incubated and examined every 24 h for 7 days. B. pseudomallei was identified by colony morphology, positive oxidase test result, inability to assimilate arabinose, antimicrobial drug susceptibility pattern (B. pseudomallei is generally resistant to gentamicin and colistin but susceptible to amoxicillin/clavulanic acid [1,2]), and results of API 20NE (bioMérieux) and B. pseudomallei–specific (Bps) latex-agglutination tests (18,24,25). Positive results were confirmed with molecular analysis. Soil type was determined by standard lithologic and pedologic analysis of sediments; for this purpose, 2 extra samples were collected per site from a depth of 30 cm (26). Sediment properties were compared with properties of other (typical) samples from the same locations as described in the recently published Soil Atlas of Africa (26).
Genetic and Phylogenetic Analyses
Genomic DNA was extracted by using a DNeasy Blood and Tissue Kit (QIAGEN, Valencia, CA, USA) to perform multilocus sequence typing (MLST) (27). Primers used to amplify fragments of the 7 housekeeping genes were identical to those described at the Burkholderia MLST website (http://bpseudomallei.mlst.net/misc/info2.asp). For isolate B. thailandensis D50, the primer narK-up was replaced by narK-upAMC 5′-TCTCTACTCGTGCGCTGGGG-3′. Sequences of the 7 gene fragments of isolates from Africa were concatenated and combined with those from a selection of 971 sequence types (STs) representing all B. pseudomallei, B. mallei, and B. thailandensis isolates in the B. pseudomallei MLST database. Concatenated sequences were aligned and analyzed by using MEGA-6 (http://www.megasoftware.net). A phylogenetic tree was constructed by using a neighbor-joining algorithm and the Kimura 2-parameter model. Bootstrap testing was performed for 500 repetitions. Whole-genome sequencing was performed by using the MiSeq platform (Illumina, San Diego, CA, USA) as described (9).
Community-Acquired Bloodstream Infections
Of the 941 bacterial blood cultures, 77 (8.2%) were positive for bacteria. The most prevalent isolate was Escherichia coli, responsible for 8 (10.0%) bloodstream infections, followed by Staphylococcus aureus (6 [7.8%]) and Salmonella enterica (6 [7.8%], 5 of which were nontyphoidal salmonellae). Other organisms that were isolated at least 5 times included Streptococcus pneumoniae (5 [6.5%]), Klebsiella pneumoniae (5 [6.5%]), and Enterobacter spp. (5 [6.5%]). B. pseudomallei was isolated from 1 (1.4%) patient, described in the case report.
Case Report
A 62-year-old Gabonese woman was hospitalized in January 2013 with a 7-day history of fever, cough, weakness, headache, vomiting, and a painful knee. She did not report coughing or shortness of breath. She had poorly controlled diabetes mellitus and was taking glibenclamide. She had no history of cardiopulmonary or renal disease, was receiving no long-term medications other than glibenclamide, and did not smoke. She was a retired school teacher but still engaged in family farming. Physical examination revealed blood pressure of 160/90 mm Hg, a pulse rate of 130 beats per minute, and a temperature of 40.5°C. She had a wound with an underlying abscess on her right leg, together with diffuse tenderness of the right knee with warmth, erythema, and limitation of active and passive ranges of motion because of pain and effusion. Neurologic, cardiovascular, and respiratory examinations revealed no abnormalities. Laboratory findings obtained at admission showed an elevated blood glucose level of 24 mmol/L but values within reference range for creatinine (0.85 mg/dL), leukocytes (9,800 × 103 mm3), and hemoglobin (9.2 g/dL). No other blood or urine test was performed, and chest radiographs were not taken. On hospitalization day 1, treatment with amoxicillin/clavulanic acid was empirically initiated for sepsis. On day 2, the abscess was incised and drained, and on day 3 antimicrobial drug therapy was switched to ceftriaxone. Cultures of blood, wound, and synovial fluid grew identical gram-negative rods, which were initially classified as Pseudomonas spp. No other pathogens were detected. The patient’s clinical condition deteriorated, and she died of septic shock on day 8. A postmortem examination was not performed.
After the patient’s death, the Pseudomonas species was classified as B. pseudomallei (patient strain Gb100) and confirmed by MLST and whole-genome sequencing. This isolate was later determined to be susceptible to trimethoprim/sulfamethoxazole, amoxicillin/clavulanic acid, ceftazidime, and meropenem (Table 1).
Seroprevalence
Of the 304 healthy schoolchildren for whom serum samples were tested for B. pseudomallei antibodies, 143 (47.0%) were male. Details for this cohort have been reported previously (15). For 43 (14.1%) children, an IHA titer was detectable; titers ranged from 1:10 to 1:80 (median 1:10, interquartile range 1:10–1:20). For 5 (1.6%) children, IHA titer was >1:40, which has been used as the cutoff value for seropositivity (22). None of the children had an IHA titer >1:160, which is considered by several centers in Thailand to support a diagnosis of melioidosis in patients with clinical features consistent with this diagnosis.
Environmental Isolates
The predominant soil type in this area of Gabon was ferralsol, which is red and yellow weathered soil. The only exception was samples taken from a rice paddy near Mouila village, where the soil was gleysol (clay, a hydric soil saturated with groundwater long enough to develop a characteristic gleyic color pattern) (Table 2). B. pseudomallei was isolated from 21 (3%) of 800 soil samples taken from 3 (38%) of the 8 sample sites; the maximum number of positive samples for 1 site was 14 (14%) (Table 2). The biochemical profiles of all isolates were in accordance with B. pseudomallei (API 20NE code 1156576). The antibiogram of B. pseudomallei soil strain C2 is shown in Table 1.
The closely related B. thailandensis coexists with B. pseudomallei in the soil in Southeast Asia and Australia and is generally considered avirulent (5,28). We also identified B. thailandensis in the soil of Gabon (Figure 2). This strain, termed B. thailandensis soil strain D50, was positive by Bps latex agglutination. This B. thailandensis strain, API 20NE code 1157577, was susceptible to trimethoprim/sulfamethoxazole, amoxicillin/clavulanic acid, ceftazidime, and meropenem (Table 1).
Genetics and Phylogeny of Burkholderia spp. Strains
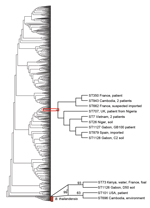
The 3 isolates from Gabon contained previously described MLST alleles but belonged to novel STs. The patient isolate Gb100 (ST1127) and soil isolate C2 (ST1128) were single-locus variants and differed by 1 nt in the narK sequence only. Patient isolate Gb100 was also a single-locus variant of ST707 (single-nucleotide substitution in ndh). The only B. pseudomallei strain with ST707 in the database had been isolated in 2010 from a patient in the United Kingdom, 6 weeks after the patient had returned from a trip to Nigeria (12). The soil isolate C2 (ST1128) was a single-locus variant of ST7 (single-nucleotide substitution in ndh) and ST879 (single-nucleotide substitution in lipA). ST7 was represented by 2 isolates in the MSLT database, both isolated in 1963 from patients in Vietnam. The B. pseudomallei ST879 strain was isolated in 2011 from a patient in Spain, who had returned from a trip through Madagascar and 14 countries in West Africa (11). The soil isolate D50 (ST1126) was a single-locus variant of ST73. This ST is represented in the database by 2 B. thailandensis strains, 1 isolated from a foal in France and 1 isolated from the environment in Kenya. Phylogenetic analysis of the Gabon isolates together with 971 STs obtained from the MLST database by using the aligned concatenated sequences of the 7 loci in the neighbor-joining algorithm with the Kimura 2-parameter model showed that the patient isolate Gb100 and soil isolate C2, found near the community of the patient, grouped together with 7 STs. These 7 STs represented 10 B. pseudomallei strains isolated in Cambodia (2 strains), Vietnam (2 strains), Niger, Nigeria, Spain (imported), France (2 strains [1 imported]), and the United Kingdom (imported) (Figure 2). Again, patient isolate Gb100 and soil isolate C2 are most closely related to ST879. Soil isolate D50 grouped together with 3 STs representing 4 B. thailandensis strains isolated from Kenya, France, the United States, and Cambodia. Using this approach, we showed that the closest relatives of the strain that infected and eventually killed the patient reported here were ST879 and the strain isolated from soil around her community. Our whole-genome sequencing sample data have been submitted to a project that is undertaking whole-genome sequencing on a large number of B. pseudomallei isolates from around the world. This approach is anticipated to offer superior resolution of the global phylogeny of B. pseudomallei (9).
We detected a case of melioidosis in a human in central Africa, confirmed the presence of B. pseudomallei in the environment in Gabon, and isolated B. thailandensis from an environmental sample from that part of the world. The low rate of antibody seropositivity among healthy children combined with the low prevalence of B. pseudomallei cultured from blood of patients in a local hospital, however, suggest that melioidosis is rare in this setting.
Only 4 of the 13 melioidosis cases acquired by humans in Africa and reported in the literature have been PCR confirmed (9–12,29–34). We show with phylogenetic analysis that the newly identified patient isolate Gb100 groups with a B. pseudomallei isolate from a patient from Spain who had traveled across West Africa and Madagascar (12). B. pseudomallei seropositivity was reported during a World Health Organization investigation into an outbreak of severe pneumonia in the northeastern of the Democratic Republic of Congo (Eric Bertherat, pers. comm.) (35). However, in that study some of the B. pseudomallei–seropositive cases diagnosed as melioidosis were later diagnosed as plague, calling into question the value of serology-based testing in this setting (35). The predominant soil type at the sites from which B. pseudomallei was isolated was similar to the soil type from which B. pseudomallei strains were isolated in Cambodia (26,36). The low rate of B. pseudomallei positivity per site points toward a relatively low abundance of B. pseudomallei in Gabon soil when compared with highly melioidosis-endemic areas in Southeast Asia and Australia (21,37). The true distribution of melioidosis in Africa remains uncertain, but we now can expand this area toward the central African country of Gabon.
The genus Burkholderia comprises >30 species, of which B. pseudomallei and B. mallei are considered the most pathogenic (2,38). The isolation of B. thailandenis from soil in Gabon extends our knowledge of the geographic distribution of this species. This strain was positive by Bps latex agglutination; this finding is in agreement with previous findings of a B. thailandensis strain from Thailand with a Bps-like capsular polysaccharide variant that also had a positive Bps latex-agglutination result (39). Our phylogenic analysis shows a divergence between the strain from Gabon and the original B. thailandenis E264 from Thailand, which is the most studied strain (4,5). Evidence of the presence of this bacterium in Africa will have implications for bacterial identification in clinical laboratories, diagnostic serology assays, and environmental studies.
Our study has several limitations. B. pseudomallei serology can be misleading; false- positive results are a major concern (40). Clearly, for assessing exposure to B. pseudomallei, an accurate, inexpensive, simple serologic assay is needed. In the interim, however, serologic evidence of exposure should be based on assays with known sensitivity and specificity against culture-confirmed melioidosis, and, to our understanding, the IHA is the best test for identifying melioidosis cases. Given the nature of working in a resource-poor environment, only limited information is available on the patient reported here (e.g., no imaging was performed to investigate the presence of deeper abscesses). With regard to the environmental study, B. pseudomallei is known for its capacity to survive in water and has been reported to be present in the air during severe weather (17); we, however did not investigate its presence in water and air in Gabon in this study. Furthermore, we cannot dismiss the possibility of error during soil sampling although guidelines for environmental sampling of B. pseudomallei were followed (17).
In summary, we identified B. pseudomallei and B. thailandensis in the Gabon environment and discovered a novel B. pseudomallei ST that can cause lethal septic shock. B. pseudomallei is probably an underrecognized cause of disease in central Africa. We propose that melioidosis occurs in central Africa but that it is unrecognized because of the lack of diagnostic microbiology facilities.
Dr. Wiersinga divides his time between patient care, teaching, and research at the Academic Medical Center, Amsterdam. His research focus is sepsis.
Acknowledgments
We thank our colleagues in the field for fruitful discussions leading towards this project, Sebastiaan Stolp for help with the logistics concerning the soil sampling study, Katja de Jong and Jacqueline Lankelma for help in the laboratory, and Matt T. Holden for help with the genetic analysis.
This study was supported by the Netherlands Organization for Scientific Research (Veni grant no. 91610008 to W.J.W.) and the Netherlands Organization for Health Research and Development (ZonMW clinical fellowship grant no. 90700424 to W.J.W.). V.W. and D.L. work at Mahidol-Oxford Tropical Medicine Research Unit funded by the Wellcome Trust of Great Britain (no. 089275/Z/09/Z).
References
- Wiersinga WJ, Currie BJ, Peacock SJ. Melioidosis. N Engl J Med. 2012;367:1035–44. DOIPubMedGoogle Scholar
- Cheng AC, Currie BJ. Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev. 2005;18:383–416. DOIPubMedGoogle Scholar
- Department of Health and Human Services, Centers for Disease Control and Prevention. Possession, use, and transfer of select agents and toxins; biennial review. Fed Regist. 2012;77:61083–115.
- Brett PJ, Deshazer D, Woods DE. Burkholderia thailandensis sp. nov., a Burkholderia pseudomallei–like species. Int J Syst Bacteriol. 1998;48:317–20. DOIPubMedGoogle Scholar
- Wiersinga WJ, de Vos AF, de Beer R. Wieland CW, Roelofs JJ, Woods DE, et al. Inflammation patterns induced by different Burkholderia species in mice. Cell Microbiol. 2008;10:81–7.
- Smith MD, Angus BJ, Wuthiekanun V, White NJ. Arabinose assimilation defines a nonvirulent biotype of Burkholderia pseudomallei. Infect Immun. 1997;65:4319–21 .PubMedGoogle Scholar
- Currie BJ, Ward L, Cheng AC. The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20 year Darwin prospective study. PLoS Negl Trop Dis. 2010;4:e900. DOIPubMedGoogle Scholar
- Rolim DB, Vilar DC, Sousa AQ. Miralles IS, de Oliveira DC, Harnett G, et al. Melioidosis, northeastern Brazil. Emerg Infect Dis. 2005;11:1458–60.
- Katangwe T, Purcell J, Bar-Zeev N. Denis B, Montgomery J, Alaerts M, et al. Human melioidosis, Malawi, 2011. Emerg Infect Dis. 2013;19:981–4.
- Cuadros J, Gil H, Miguel JD. Marabé G, Gómez-Herruz TA, Lobo B, et al. Case report: melioidosis imported from West Africa to Europe. Am J Trop Med Hyg. 2011;85:282–4.
- Morosini MI, Quereda C, Gil H. Anda P, Núñez-Murga M, Cantón R, et al. Melioidosis in traveler from Africa to Spain. Emerg Infect Dis. 2013;19:1656–9.
- Salam AP, Khan N, Malnick H, Kenna DT, Dance DA, Klein JL. Melioidosis acquired by traveler to Nigeria. Emerg Infect Dis. 2011;17:1296–8. DOIPubMedGoogle Scholar
- Batchelor BI, Paul J, Trakulsomboon S, Mgongo M, Dance DA. Melioidosis survey in Kenya. Trans R Soc Trop Med Hyg. 1994;88:181. DOIPubMedGoogle Scholar
- Currie BJ, Dance DA, Cheng AC. The global distribution of Burkholderia pseudomallei and melioidosis: an update. Trans R Soc Trop Med Hyg. 2008;102:S1–4. DOIPubMedGoogle Scholar
- Lell B, Faucher JF, Missinou MA. Borrmann S, Dangelmaier O, Horton J, et al. Malaria chemoprophylaxis with tafenoquine: a randomised study. Lancet. 2000;355:2041–5.
- Suputtamongkol Y, Hall AJ, Dance DA. Chaowagul W, Rajchanuvong A, Smith MD, et al. The epidemiology of melioidosis in Ubon Ratchatani, northeast Thailand. Int J Epidemiol. 1994;23:1082–90.
- Wuthiekanun V, Chantratita N, Dance D, Limmathurotsakul D, Peacock SJ. SOP: latex agglutination technique for the detection of Burkholderia pseudomallei. 2012 [cited 2014 May 1]. http://www.melioidosis.info/download.aspx
- Jenney AW, Lum G, Fisher DA, Currie BJ. Antibiotic susceptibility of Burkholderia pseudomallei from tropical northern Australia and implications for therapy of melioidosis. Int J Antimicrob Agents. 2001;17:109–13. DOIPubMedGoogle Scholar
- Alexander AD, Huxsoll DL, Warner AR Jr, Shepler V, Dorsey A. Serological diagnosis of human melioidosis with indirect hemagglutination and complement fixation tests. Appl Microbiol. 1970;20:825–33 .PubMedGoogle Scholar
- Cheng AC, O'Brien M, Freeman K, Lum G, Currie BJ. Indirect hemagglutination assay in patients with melioidosis in northern Australia. Am J Trop Med Hyg. 2006;74:330–4 .PubMedGoogle Scholar
- Limmathurotsakul D, Wuthiekanun V, Chantratita N. Wongsuvan G, Amornchai P, Day NP, et al. Burkholderia pseudomallei is spatially distributed in soil in northeast Thailand. PLoS Negl Trop Dis. 2010;4:e694.
- Wuthiekanun V, Anuntagool N, White NJ, Sirisinha S. Short report: a rapid method for the differentiation of Burkholderia pseudomallei and Burkholderia thailandensis. Am J Trop Med Hyg. 2002;66:759–61 .PubMedGoogle Scholar
- Amornchai P, Chierakul W, Wuthiekanun V. Mahakhunkijcharoen Y, Phetsouvanh R, Currie BJ, et al. Accuracy of Burkholderia pseudomallei identification using the API 20NE system and a latex agglutination test. J Clin Microbiol. 2007;45:3774–6.
- Jones A, Breuning-Madson H, Brossard M. Soil atlas of Africa. European Commission. Luxembourg (Belgium): Publications Office of the European Union; 2013.
- Godoy D, Randle G, Simpson AJ. Aanensen DM, Pitt TL, Kinoshita R, et al. Multilocus sequence typing and evolutionary relationships among the causative agents of melioidosis and glanders, Burkholderia pseudomallei and Burkholderia mallei. J Clin Microbiol. 2003;41:2068–79.
- Brett PJ, Deshazer D, Woods DE. Characterization of Burkholderia pseudomallei and Burkholderia pseudomallei–like strains. Epidemiol Infect. 1997;118:137–48. DOIPubMedGoogle Scholar
- Bremmelgaard A, Bygbjerg I, Hoiby N. Microbiological and immunological studies in a case of human melioidosis diagnosed in Denmark. Scand J Infect Dis. 1982;14:271–5 .PubMedGoogle Scholar
- Wall RA, Mabey DC, Corrah PT, Peters L. A case of melioidosis in West Africa. J Infect Dis. 1985;152:424–5. DOIPubMedGoogle Scholar
- Issack MI, Bundhun CD, Gokhool H. Melioidosis in Mauritius. Emerg Infect Dis. 2005;11:139–40. DOIPubMedGoogle Scholar
- Martinet O, Pac Soo AM, Knezynski M. Melioidosis: regarding a case acquired in Madagascar and two nosocomial cases [in French]. Bull Soc Pathol Exot. 2004;97:366–70.
- Borgherini G, Poubeau P, Paganin F. Picot S, Michault A, Thibault F, et al. Melioidosis: an imported case from Madagascar. J Travel Med. 2006;13:318–20.
- Amezyane T, Lecoules S, Algayres JP. Mycotic iliac aneurysm associated with Burkholderia pseudomallei. Int J Infect Dis. 2010;14(Suppl 3):e381–2. DOIPubMedGoogle Scholar
- Rattanavong S, Wuthiekanun V, Langla S. Amornchai P, Sirisouk J, Phetsouvanh R, et al. Randomized soil survey of the distribution of Burkholderia pseudomallei in rice fields in Laos. Appl Environ Microbiol. 2011;77:532–6.
- Ussery DW, Kiil K, Lagesen K, Sicheritz-Ponten T, Bohlin J, Wassenaar TM. The genus Burkholderia: analysis of 56 genomic sequences. Genome Dyn. 2009;6:140–57. DOIPubMedGoogle Scholar
- Hantrakun V, Rongkard P, Amonchi P, Sarunporn T, Langla S, Wuthiekanun V, Presence of environmental B. pseudomallei, B. thailandensis and putative B. thailandensis with B. ps–like CPS variant in east Thailand. Abstract in: Proceedings of the 7th World Melioidosis Congress; 2013 Sep 18–20; Bangkok, Thailand.
- Peacock SJ, Cheng AC, Currie BJ, Dance DA. The use of positive serological tests as evidence of exposure to Burkholderia pseudomallei. Am J Trop Med Hyg. 2011;84:1021–2. DOIPubMedGoogle Scholar
Figures
Tables
Cite This Article1These authors contributed equally to this article.
Table of Contents – Volume 21, Number 1—January 2015
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
W. Joost Wiersinga, Center for Infection and Immunity Amsterdam, Dept. of Internal Medicine, Academic Medical Center, University of Amsterdam, Meibergdreef 9, Room G2-132, 1105 AZ Amsterdam, the Netherlands
Top