Volume 21, Number 4—April 2015
Letter
Enterovirus A71 Subgenotype B5, France, 2013
Figure
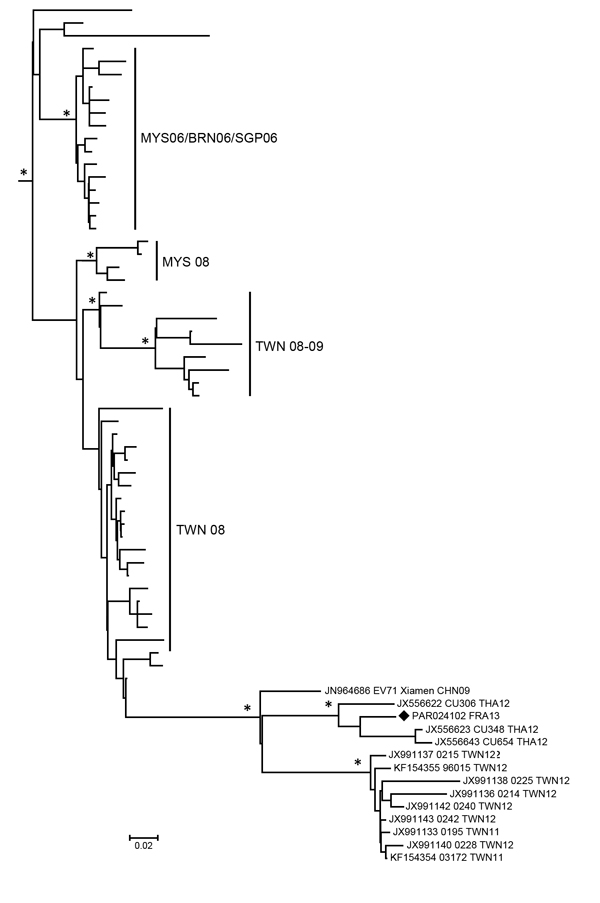
Figure. Phylogeny of enterovirus A71 (EV-A71) subgenogroups B4 and B5 inferred with 274 partial 1D gene sequences, France. Black diamond indicates strain PAR024103_FRA13 from this study. The phylogenetic relationships were inferred following a Bayesian method by using a relaxed molecular clock model with an uncorrelated exponential distribution of evolution rates estimated with a general time reversible substitution model and a Bayesian skyline plot as a population model (BEAST version 1.7.5; http://beast.bio.ed.ac.uk/). Sequences were 319 bp. For clarity, only a subtree of EV-A71 B5 sequences is shown, and taxon names are not included. Asterisks indicate nodes with posterior probability density values >0.90. Geographic origins and time of isolation of strains are indicated by the International Organization of Standardization code abbreviation followed by the year of isolation. Scale bar indicates nucleotide substitutions per site.