Volume 21, Number 7—July 2015
Dispatch
Hemagglutinin Receptor Binding of a Human Isolate of Influenza A(H10N8) Virus
Abstract
Three cases of influenza A(H10N8) virus infection in humans have been reported; 2 of these infected persons died. Characterization of the receptor binding pattern of H10 hemagglutinin from avian and human isolates showed that both interact weakly with human-like receptors and maintain strong affinity for avian-like receptors.
Human infections with avian influenza A(H10N8) virus were reported in China during the 2013–14 winter influenza season. The first patient, a 73-year old woman, became ill in November 2013 a few days after visiting a live poultry market in Jiangxi Province (1). Two additional patients, a 55-year-old woman and a 75-year-old man, were admitted to hospitals in the same province in January 2014 (2). Severe pneumonia and subsequent acute respiratory distress syndrome developed in all 3 patients; 2 of the patients died, 5 and 6 days after admission (2).
Epithelial cells of the human upper respiratory tract contain mostly α2,6-linked sialic acids (SAα2,6) and low levels of α2,3-linked sialic acids (SAα2,3) (3). Hemagglutinin (HA) of avian influenza virus strains shows preferential binding to SAα2,3 receptors, which partially accounts for the reduced ability of avian influenza strains to establish infections in humans (3). Interaction with SAα2,6 receptors is one of the requirements for efficient replication in the human upper respiratory tract. In addition, reduced binding to SAα2,3 facilitates respiratory droplet-based transmission in ferrets (4). Therefore, emerging avian influenza viruses with increased binding to SAα2,6 and reduced binding to SAα2,3 pose a major pandemic threat, and active research and surveillance to detect animal viruses with modified receptor binding are warranted.
We analyzed the amino acid sequence of the receptor binding site of HA from the isolate A/Jiangxi-Donghu/346-1/2013 (H10-JD346; Global Initiative on Sharing Avian Influenza Data [GISAID, http://www.gisaid.org] accession no. EPI530526) from the first patient infected by influenza A(H10N8) virus. In addition, several human and avian influenza viruses (sequences from GISAID or the National Center for Biotechnology Information website) and a recent harbor seal isolate (5) were compared with H10-JD346 (Table). We observed that residues involved in receptor binding for H10 subtype influenza viruses suggest avian-like receptor specificity. However, we identified 2 amino acids in avian and human H10, T135 and S186, that are common in circulating human influenza viruses and were associated with changes in receptor binding in other avian influenza A virus subtypes (6,7). In accordance with this finding, Vachieri et al. found substantial levels of binding of an avian H10 HA to SAα2,6 that retained the ability to interact with SAα2,3 (8).
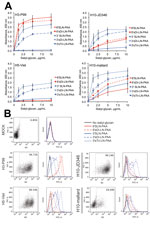
Given the role of receptor binding specificity of emerging influenza viruses, we analyzed the interaction of HA of the human H10-JD346 influenza A(H10N8) virus isolate in comparison with that of an avian H10N7 subtype virus. First, we used a solid-phase binding assay (9,10) and the following biotinylated glycans conjugated with a polyacrylamide (PAA) support (provided by the Consortium of Functional Glycomics [CFG]): Neu5Acα2,6Galβ1–4GlcNAcβ-PAA (6′ SLN-PAA); Neu5Acα2–6(Galβ1–4GlcNAcβ1–3)2β-PAA (6’sDi-LN-PAA); Neu5Acα2,3Galβ1–4GlcNAcβ-PAA (3′ SLN-PAA); Neu5Acα2–3(Galβ1–4GlcNAcβ1–3)2β-PAA (3′sDi-LN-PAA); and Neu5Acα2–3(Galβ1–4GlcNAcβ-sp)3β-PAA (3′sTri-LN-PAA). We also analyzed recombinant hexahistidine-tagged HAs (11) from H10-JD346, an avian H10N7 subtype strain from North America (A/mallard/Interior Alaska/10BM01929/2010; H10-mallard), a human H3N2 subtype seasonal influenza A virus (A/Panama/2007/1999; H3-P99), and an H5N1 subtype avian influenza virus from a fatal human case (A/Vietnam/1203/2004; H5-Viet).
As expected, H3-P99 bound strongly to the SAα2,6 tested, and H5 showed higher levels of binding to SAα2,3 than to SAα2,6 (Figure 1, panel A). When we analyzed H10-mallard and H10-JD346, we found a similar binding profile, which is consistent with the presence of similar amino acids affecting the receptor binding specificity (Table). Although both H10 proteins had a prevalent avian-like binding profile, low levels of binding to SAα2,6 were also observed.
To confirm this data, we used a flow cytometry–based assay and the same synthetic glycans (9,10). We infected MDCK epithelial cells with H10-JD346 virus (6:2 re-assortant with the backbone of laboratory strain A/Puerto Rico/8/1934 [PR8], which was generated as described) (9,10); H10-mallard (wild-type); human isolate H3-P99 (wild-type); and H5-Viet 6:2 (low pathogenicity reassortant with the backbone of PR8) (9,10) at a multiplicity of infection of 1. Cells were harvested 24-h postinfection and incubated with antibody against matrix protein 2 (E10), which was detected by using an antibody against IgG (Alexa 647 antibody; Invitrogen, Carlsbad, CA, USA) as a control of infection and with the sialyl-glycans (detected with streptavidin–fluorescein isothiocyanate; Jackson Laboratories, Bar Harbor, ME, USA). We determined the percentage of infected cells in each sample and gated the infected population to determine the SA binding profile (Figure 1, panel B).
H3-P99 showed high levels of binding to SAα2,6 and H5-Viet bound more efficiently to SAα2,3 than to SAα2,6, which is similar to observations with recombinant HAs in the solid-phase binding assay. H10-mallard and H10-JD346 showed similar binding profiles with preferential binding for SAα2,3 and binding to SAα2,6 slightly higher than that for the negative control.
Analysis of receptor binding of H10-JD346 and of H10-mallard with 2 independent assays indicated that the H10 subtype influenza virus interacts slightly with human-like receptors and maintains preferential binding to avian-like receptors. Consequently, these data suggest that H10 subtype influenza virus might have the ability to interact with the upper human respiratory tract, which is rich in SAα2,6 (3).
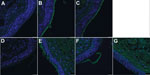
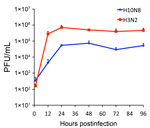
To test this hypothesis, we precomplexed H3-P99 and H10-JD346 with primary antibody (mouse anti-His tag) and secondary fluorescent antibody, then incubated the complex with 2 human tracheal samples (12). As expected, H3-P99 HA bound to the surface of respiratory epithelia (Figure 2). Recombinant H10-JD346 HA also interacted with respiratory epithelia (Figure 2), which suggested that the virus might be able to attach and replicate in the human upper respiratory tract. However, the 6:2 reassortant virus H10-JD346 virus showed markedly decreased replication compared with that of an H3N2 subtype virus (PR8 6:2 reassortant) in a human lung epithelial cell line (Figure 3).
HA of novel influenza A(H10N8) virus interacts with SAα2,3 and slightly with SAα2,6, at levels similar to that for an avian H10 subtype HA, and binds to cells in the human upper respiratory tract. Our findings are consistent with those of Vachiery et al. (8) but show some differences from those of Yang et al. (13) and Wang et al. (14), who did not detect interaction with SAα2,6 or human trachea. Variations in the experimental settings and protocols (e.g., concentration of HA or glycans used) might account for these dissimilarities.
Only 3 cases of human infections with influenza A(H10N8) viruses have been reported. However, H10N7 subtype viruses have caused conjunctivitis or mild respiratory symptoms in humans. An epidemic among seals caused by this virus subtype is currently ongoing in Europe (5). A study by Beare and Webster showed that ≈50% of volunteers experimentally infected with influenza A(H10N7) virus shed virus (15), which our data suggests might be caused by initial attachment to the upper respiratory tract. Immune responses were not detected in these volunteers, and mild, if any, symptoms developed, which indicated limited virus replication.
The low incidence of H10 influenza virus indicates a limited pandemic potential of H10N7 and H10N8 viruses. Therefore, further changes in receptor binding, as well as acquisition of genomic segments from other avian influenza virus strains through co-infection, would be required to increase fitness and transmissibility in mammals. Isolate H10-JD346 amino acid sequence had a mixture of E and K in position 627 of basic polymerase protein 2; the K627 mutation is associated with mammal adaptation (1). This finding highlights the need for an efficient surveillance network to track and identify possible changes, as well as extensive research to identify them and understand their functional consequences.
Dr. Ramos is an assistant professor in the Department of Microbiology at the Icahn School of Medicine at Mount Sinai, New York, NY. Her main research interests focus on receptors for influenza viruses and innate immunity to these viruses.
Acknowledgments
We thank CFG for providing reagents. Some of the data will be published on the CFG website (http://www.functionalglycomics.org/). We also thank the Flow Cytometry Shared Facility and the Microscopy Core Facility at Icahn School of Medicine at Mount Sinai for assistance, the Icahn School of Medicine Institutional Biorepository for providing human tissue sections, John Steel and Randy A. Albrecht for providing recombinant influenza viruses, and GISAID for making H10N7 and H10N8 subtype virus sequencing data publicly available.
This study was supported by the National Institutes of Health/National Institute of Allergy and Infectious Diseases (Center for Research on Influenza Pathogenesis, contract HHSN272201400008C to J.A.R., F.K., and A.F.-S.) as part of the Centers for Excellence for Influenza Research and Surveillance Network. I.R. was partially supported by National Institutes of Health training grant T32 AI788926.
References
- Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. Lancet. 2014;383:714–21 . DOIPubMedGoogle Scholar
- Zhang W, Wan J, Qian K, Liu X, Xiao Z, Sun J, Clinical characteristics of human infection with a novel avian-origin influenza A(H10N8) virus. Chin Med J (Engl). 2014;127:3238–42 .PubMedGoogle Scholar
- Neumann G, Kawaoka Y. Host range restriction and pathogenicity in the context of influenza pandemic. Emerg Infect Dis. 2006;12:881–6 . DOIGoogle Scholar
- Tumpey TM, Maines TR, Van Hoeven N, Glaser L, Solorzano A, Pappas C, A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science. 2007;315:655–9 . DOIGoogle Scholar
- Zohari S, Neimanis A, Harkonen T, Moraeus C, Valarcher JF. Avian influenza A(H10N7) virus involvement in mass mortality of harbour seals (Phoca vitulina) in Sweden, March through October 2014. Euro Surveill. 2014;19:20967 .PubMedGoogle Scholar
- Watanabe T, Kiso M, Fukuyama S, Nakajima N, Imai M, Yamada S, Characterization of H7N9 influenza A viruses isolated from humans. Nature. 2013;501:551–5 . DOIGoogle Scholar
- Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–69 . DOIPubMedGoogle Scholar
- Vachieri SG, Xiong X, Collins PJ, Walker PA, Martin SR, Haire LF, Receptor binding by H10 influenza viruses. Nature. 2014;511:475–7 . DOIPubMedGoogle Scholar
- Ramos I, Bernal-Rubio D, Durham N, Belicha-Villanueva A, Lowen AC, Steel J, Effects of receptor binding specificity of avian influenza virus on the human innate immune response. J Virol. 2011;85:4421–31 . DOIPubMedGoogle Scholar
- Ramos I, Krammer F, Hai R, Aguilera D, Bernal-Rubio D, Steel J, H7N9 influenza viruses interact preferentially with alpha2,3-linked sialic acids and bind weakly to alpha2,6-linked sialic acids. J Gen Virol. 2013;94:2417–23 . DOIPubMedGoogle Scholar
- Krammer F, Margine I, Tan GS, Pica N, Krause JC, Palese P. A carboxy-terminal trimerization domain stabilizes conformational epitopes on the stalk domain of soluble recombinant hemagglutinin substrates. PLoS ONE. 2012;7:e43603 . DOIPubMedGoogle Scholar
- Tharakaraman K, Jayaraman A, Raman R, Viswanathan K, Stebbins NW, Johnson D, Glycan receptor binding of the influenza A virus H7N9 hemagglutinin. Cell. 2013;153:1486–93 . DOIGoogle Scholar
- Yang H, Carney PJ, Chang JC, Villanueva JM, Stevens J. Structure and receptor binding preferences of recombinant hemagglutinins from avian and human H6 and H10 influenza A virus subtypes. J Virol. 2015 Feb 11;pii: JVI.03456-14.PubMedGoogle Scholar
- Wang M, Zhang W, Qi J, Wang F, Zhou J, Bi Y, Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus. Nat Commun. 2015;6:5600.PubMedGoogle Scholar
- Beare AS, Webster RG. Replication of avian influenza viruses in humans. Arch Virol. 1991;119:37–42 . DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleTable of Contents – Volume 21, Number 7—July 2015
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Irene Ramos, Department of Microbiology, Icahn School of Medicine at Mount Sinai, 1468 Madison Ave, Box 1124, New York, NY 10029, USA; ; or Florian Krammer, Department of Microbiology, Icahn School of Medicine at Mount Sinai, 1 Gustave L. Levy Pl, Box 1124, New York, NY 10029, USA
Top