Volume 22, Number 12—December 2016
Dispatch
Detection of Vaccinia Virus in Dairy Cattle Serum Samples from 2009, Uruguay
Abstract
We detected orthopoxvirus in 28 of 125 serum samples collected during 2009 from cattle in Uruguay. Two samples were PCR-positive for vaccinia virus and had sequences similar to those for vaccinia virus associated with outbreaks in Brazil. Autochthonous circulation of vaccinia virus in Uruguay and other South American countries cannot be ruled out.
Orthopoxviruses (family Poxviridae, genus Orthopoxvirus) cause several zoonotic diseases worldwide, including diseases caused by monkeypox virus in Africa, cowpox virus mainly in Europe, and vaccinia virus (VACV) in South America and Asia (1). During the past few decades, reports about emerging and reemerging zoonotic VACV and buffalopox virus have increased, as have the number of severe cases of disease caused by the viruses (2,3). VACV has been isolated in Brazil and detected in Argentina (4–6). During bovine vaccinia outbreaks, VACV affects mainly dairy herds; lesions develop on the animals, especially on the teats and udders, resulting in reduced milk production (5,6). In humans, most VACV infections occur in persons who milk cattle; infection frequently causes lesions on hands and forearms, but systemic clinical manifestations have been described and represent a challenge to public health services (5).
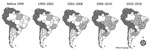
The first notifications of VACV detection in Brazil were in the 1960s and 1970s during a government surveillance campaign that investigated emerging pathogens in wild animals (5). However, it was not until 1999 that the first outbreaks of bovine vaccinia were reported in Brazil, when cases occurred in Rio de Janeiro and São Paulo States (5). Over the next few years, VACV spread to several more states; since then, all geographic regions of Brazil have been affected by bovine vaccinia, including states bordering other countries in South America, which explains the recent detection of VACV in Argentina (Figure 1) (4–8). Given these detections, serious concern exists regarding the potential spread of VACV to other countries in South America. Uruguay, a country that borders Brazil, has had no reports of VACV detection. To determine if the virus has spread to Uruguay, we investigated the presence of orthopoxvirus neutralizing antibodies and viral DNA in serum samples from cattle in the country.
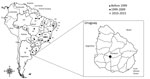
We analyzed serum samples that were collected in May 2009 from 125 dairy cows in Durazno County (33°23′0′′S, 56°31′0′′W), Durazno State, Uruguay (Figure 2). The cattle herds had no clinical sings of disease at the time of serum collection. To determine the presence of neutralizing antibodies in the serum samples, we used an orthopoxvirus plaque-reduction neutralization test as previously described (9). The serum titer was defined as the highest dilution that inhibited >70% of virus plaques compared with negative controls (4).
Because previous studies have detected viral DNA in the serum of animals and humans with and without clinical manifestations (4,10,11), we performed a molecular investigation to identify orthopoxvirus. We used quantitative PCR (qPCR) to amplify VACV growth factor gene (C11R) DNA. This qPCR tool has high sensitivity and specificity and, thus, has been routinely used as an orthopoxvirus diagnostic tool by our group (12). For molecular characterization, we used the nonconserved orthopoxvirus hemagglutinin gene (A56R) (13). We used VACV–Western Reserve as the PCR-positive control for amplification and characterization.
The PCR A56R products obtained from C11R PCR–positive samples were sequenced in both orientations and subjected to capillary electrophoresis (3130 Genetic Analyzer, BigDye Terminator Cycle Sequencing Kit v3.1; Applied Biosystems, Foster City, CA, USA). We used the ClustalW (http://www.clustal.org/) method to align sequences with previously published orthopoxvirus sequences from GenBank; alignments were manually checked with MEGA6 (http://www.megasoftware.net/). We constructed phylogenetic trees using the neighbor-joining method with 1,000 bootstrap replicates and the Tamura 3-parameter model in MEGA6. All field and laboratory clinical samples were processed separately to avoid cross-contamination. Serologic and molecular tests were performed in 2 independent experiments and in duplicate.
We detected neutralizing antibodies against orthopoxvirus in 28 (22.4%) of 125 serum samples from cattle in Uruguay; titers were 100 neutralizing units (NU)/mL in 10 (35.7%) samples, 200 NU/mL in 11 (39.3%) samples, 400 NU/mL in 5 (17.9%) samples, and 800 NU/mL in 2 (7.1%) samples (Technical Appendix Figure 1, panel A). To confirm that the orthopoxvirus seropositivity represented seropositivity to VACV, we used the C11R gene to molecularly test DNA in the serum samples; 2 (1.6%) samples were positive by qPCR, and 1 of those was also positive by the plaque-reduction neutralization test.
One of the 2 C11R PCR–positive isolates was also positive for the A56R gene; we sequenced the gene and named the strain VACV Uruguay. Alignment of the A56R nucleotide sequence showed the presence of an 18-nt signature deletion, which is also present in sequences of Brazilian-VACV group I, but not group II, viruses. Unlike sequences for other VACVs, the sequence for VACV Uruguay had an A→T polymorphism (Technical Appendix Figure 2, panel A). VACV Uruguay exhibited higher identity with group I (98.6% identity) viruses from Brazil and Argentina than to group II (97.3% identity) viruses from Brazil (Technical Appendix Figure 1, panel B). Furthermore, in the phylogenetic tree based on A56R nucleotide sequences, the VACVs from Uruguay clustered with group I VACVs that had been detected during outbreaks in Brazil and with viruses from Argentina (Technical Appendix Figure 2, panel B).
Since 1999, VACV has been isolated from symptomatic and asymptomatic cattle, humans, and wildlife from the north to the extreme south of Brazil (5,6,8), and in 2014, VACV was described in bovine serum samples from Argentina (4). Although no exanthematous VACV outbreaks have been reported among cattle in Uruguay, we detected orthopoxvirus antibodies and VACV DNA in serum samples from dairy cattle in the country, indicating they have been exposed to VACV. Uruguay shares a border with Brazil, and its western border is shared with Entre Rios Province in Argentina, where VACV DNA has been detected. In addition, Uruguay shares its northern and eastern borders with Rio Grande do Sul State, Brazil, where Pelotas VACV has been isolated from horses (4,5).
Our finding of orthopoxvirus antibodies and VACV DNA indicates a possible undetected or silent circulation of VACV in Uruguay. Considering the importance of the livestock sector in all countries of South America, concern exists about the possible spread of VACV beyond Brazil, Argentina, and Uruguay (4–6,14). Despite surveillance by veterinarians, efforts to stop the spread of VACV at borders may be hampered by the movement of infected rural workers, the marketing of asymptomatic live animals, and the misdiagnosis of VACV infection. Furthermore, VACV has been shown to circulate in wild environments, and it has been hypothesized that rodents may serve as VACV hosts, as they do for other orthopoxviruses, and facilitate the spread of VACV in border areas (15).
Bovine vaccinia outbreaks in South America were first reported in Brazil, but we cannot rule out the possibility of autochthonous circulation of VACV in Uruguay and other countries in South America. Additional studies are needed to elucidate VACV seroprevalence in other countries in South America, and further research is needed to clarify the transmission pathways related to the spread of VACV in South America.
Dr. Franco-Luiz is a biomedical doctor in the Laboratório de Vírus, Departamento de Microbiologia, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais. Her research focuses on the monitoring, control, and prevention of vaccinia virus infection.
Acknowledgments
We thank colleagues from the Laboratório de Vírus, Universidade Federal de Minas Gerais, for their excellent technical support.
Financial support was provided by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq); Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES); Fundação de Amparo à Pesquisa do Estado de Minas Gerais; and Ministério da Agricultura, Pecuária e Abastecimento.
A.P.M.F.L. received fellowships from CAPES. E.G.K., C.A. B., G.S.T., P.C.P. F., and J.S.A. received research fellowships from CNPq.
References
- Shchelkunov SN. An increasing danger of zoonotic orthopoxvirus infections. PLoS Pathog. 2013;9:e1003756.DOIPubMedGoogle Scholar
- Abrahão JS, Campos RK, Trindade GS, Guimarães da Fonseca F, Ferreira PC, Kroon EG. Outbreak of severe zoonotic vaccinia virus infection, Southeastern Brazil. Emerg Infect Dis. 2015;21:695–8.DOIPubMedGoogle Scholar
- Goyal T, Varshney A, Bakshi SK, Barua S, Bera BC, Singh RK. Buffalo pox outbreak with atypical features: a word of caution and need for early intervention! Int J Dermatol. 2013;52:1224–30.PubMedGoogle Scholar
- Franco-Luiz APM, Fagundes-Pereira A, Costa GB, Alves PA, Oliveira DB, Bonjardim CA, et al. Spread of vaccinia virus to cattle herds, Argentina, 2011. [letter]. Emerg Infect Dis. 2014;20:1576–8.DOIPubMedGoogle Scholar
- Kroon EG, Mota BE, Abrahão JS, da Fonseca FG, de Souza Trindade G. Zoonotic Brazilian Vaccinia virus: from field to therapy. Antiviral Res. 2011;92:150–63.DOIPubMedGoogle Scholar
- Medaglia ML, Pessoa LC, Sales ER, Freitas TR, Damaso CR. Spread of cantagalo virus to northern Brazil. Emerg Infect Dis. 2009;15:1142–3.DOIPubMedGoogle Scholar
- Damon IK. Poxviruses. In: Knipe DM, Howley PM, editors. Fields virology. 6th ed. Vol II. Philadelphia: Lippincott Williams & Wilkins; 2013. p. 2160–84.
- de Sant’Ana FJ, Leal FA, Rabelo RE, Vulcani VA, Moreira CA Jr, Cargnelutti JF, et al. Coinfection by Vaccinia virus and an Orf virus-like parapoxvirus in an outbreak of vesicular disease in dairy cows in midwestern Brazil. J Vet Diagn Invest. 2013;25:267–72.DOIPubMedGoogle Scholar
- Silva-Fernandes AT, Travassos CE, Ferreira JM, Abrahão JS, Rocha ES, Viana-Ferreira F, et al. Natural human infections with Vaccinia virus during bovine vaccinia outbreaks. J Clin Virol. 2009;44:308–13.DOIPubMedGoogle Scholar
- Cohen JI, Hohman P, Preuss JC, Li L, Fischer SH, Fedorko DP. Detection of vaccinia virus DNA, but not infectious virus, in the blood of smallpox vaccine recipients. Vaccine. 2007;25:4571–4.DOIPubMedGoogle Scholar
- Savona MR, Dela Cruz WP, Jones MS, Thornton JA, Xia D, Hadfield TL, et al. Detection of vaccinia DNA in the blood following smallpox vaccination. JAMA. 2006;295:1898–900.DOIPubMedGoogle Scholar
- Kroon E, Santos Abrahão J, de Souza Trindade G, Pereira Oliveira G, Moreira Franco Luiz AP, Barbosa Costa G, et al. Natural vaccinia virus infection: diagnosis, isolation, and characterization. Curr Protoc Microbiol. 2016;42:1, 43.
- de Souza Trindade G, Li Y, Olson VA, Emerson G, Regnery RL, da Fonseca FG, et al. Real-time PCR assay to identify variants of Vaccinia virus: implications for the diagnosis of bovine vaccinia in Brazil. J Virol Methods. 2008;152:63–71.DOIPubMedGoogle Scholar
- Hutter S, Aviles MM, Matus MCR, Maresca R. Monitoring assessment report of the veterinary services in Uruguay [in Spanish]. Organización Mundial de Sanidad Animal. 2014 [cited 2016 Jan 15]. http://www.oie.int/fileadmin/Home/eng/Support_to_OIE_Members/pdf/PVS-Final-Report-Uruguay-FU.pdf
- Abrahão JS, Guedes MI, Trindade GS, Fonseca FG, Campos RK, Mota BF, et al. One more piece in the VACV ecological puzzle: could peridomestic rodents be the link between wildlife and bovine vaccinia outbreaks in Brazil? PLoS One. 2009;4:e7428.DOIPubMedGoogle Scholar
Figures
Cite This ArticleTable of Contents – Volume 22, Number 12—December 2016
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Erna Geessien Kroon, Laboratório de Vírus, Departamento de Microbiologia, Instituto de Ciências Biológicas, Universidade Federal de Minas Gerais, Av Antônio Carlos, 6627, Caixa Postal 486, CEP 31270-901, Belo Horizonte, Minas Gerais, Brazil
Top