African Horse Sickness Caused by Genome Reassortment and Reversion to Virulence of Live, Attenuated Vaccine Viruses, South Africa, 2004–2014
Camilla Weyer, John D. Grewar, Phillippa Burger, Esthea Rossouw, Carina Lourens, Christopher Joone, Misha le Grange, Peter Coetzee, Estelle H. Venter, Darren P. Martin, N. James MacLachlan, and Alan J. Guthrie

Author affiliations: University of Pretoria, Onderstepoort, South Africa (C.T. Weyer, P. Burger, C. Lourens, C. Joone, M. le Grange, P. Coetzee, E. Venter, N.J. MacLachlan, A.J. Guthrie); Western Cape Department of Agriculture, Elsenburg, South Africa (J.D. Grewar); Wits Health Consortium, Johannesburg, South Africa (E. Rossouw); University of Cape Town, Cape Town, South Africa (D.P. Martin); University of California, Davis, CA, USA (N.J. MacLachlan)
Main Article
Figure 3
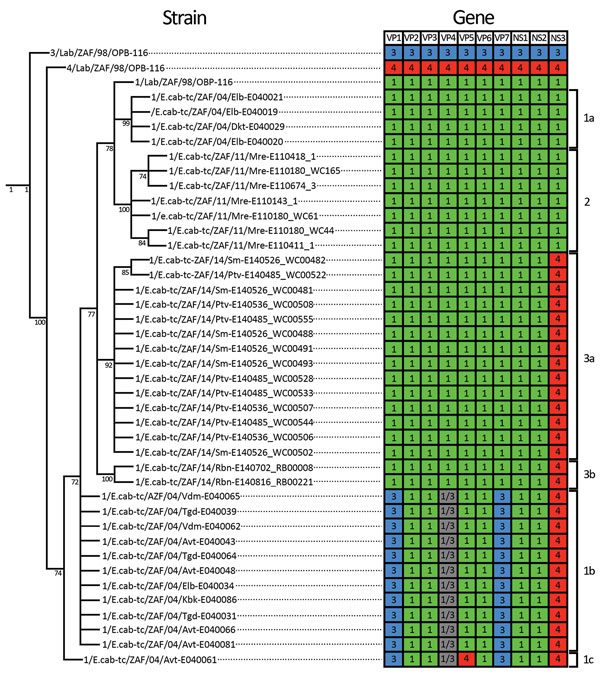
Figure 3. Cladogram and heat map of vaccine-derived African horse sickness (AHS) virus reassortants identified in AHS outbreaks in Western Cape Province, South Africa, 2004–2014. Cladogram indicates genetic relationships of concatenated AHS virus whole-genome nucleotide sequences from affected horses in the 2004, 2011, and 2014 outbreaks in the AHS controlled area in Western Cape Province. Heat map diagram summarizes the origin of the gene segments for each strain with 1/Lab/ZAF/98/OBP-116 (green blocks), 3/Lab/ZAF/98/OBP-116 (blue blocks), and 4/Lab/ZAF/98/OBP-116 (red blocks) vaccine-derived strains. Gray blocks indicate that the segment could be derived from either 1/Lab/ZAF/98/OBP-116 or 3/Lab/ZAF/98/OBP-116. Branches supported by full maximum-likelihood bootstrap values >70% are indicated. Genotype groups are indicated at right.
Main Article
Page created: November 17, 2016
Page updated: November 17, 2016
Page reviewed: November 17, 2016
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
