Volume 22, Number 3—March 2016
Dispatch
Far East Scarlet-Like Fever Caused by a Few Related Genotypes of Yersinia pseudotuberculosis, Russia
Figure 2
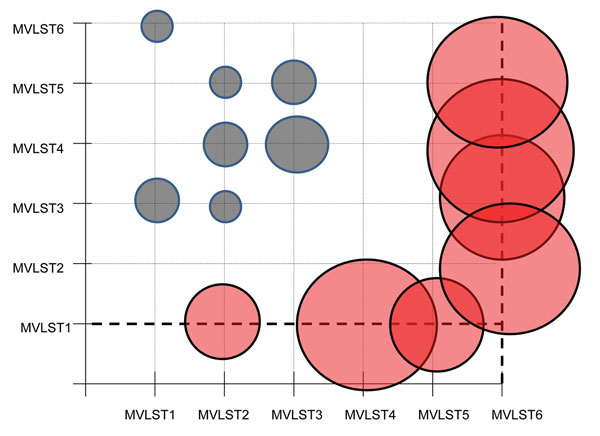
Figure 2. Graphic representation of the evolutionary analysis that tested the hypothesis of equality of evolutionary rates between multivirulence locus sequence type (MVLST) genotypes for study of Far East scarlet-like fever caused by a clonal group of Yersinia pseudotuberculosis, Russia. The χ2 test statistic was applied for the pairwise comparison of concatenated sequences of MVLST markers, with the Y. pestis sequence being used as an outgroup. Circles indicate values of the χ2 test statistic of the pairwise comparison calculated in MEGA6 (10); diameters correspond to values of rejection of the null hypothesis that states the equality of evolutionary rates between pairs of concatenated sequences. Statistically significant values are shown in red.
References
- Grunin II, Somov GP, Zalmover IIu. Far Eastern scarlatinoid fever [in Russian]. Voen Med Zh. 1960;
8 :62–6.PubMedGoogle Scholar - Fukushima H, Matsuda Y, Seki R, Tsubokura M, Takeda N, Shubin FN, Geographical heterogeneity between far eastern and western countries in prevalence of the virulence plasmid, the superantigen Yersinia pseudotuberculosis-derived mitogen, and the high-pathogenicity island among Yersinia pseudotuberculosis strains. J Clin Microbiol. 2001;39:3541–7. DOIPubMedGoogle Scholar
- Eppinger M, Rosovitz MJ, Fricke WF, Rasko DA, Kokorina G, Fayolle C, The complete genome sequence of Yersinia pseudotuberculosis IP31758, the causative agent of Far East scarlet-like fever. PLoS Genet. 2007;3:e142. DOIPubMedGoogle Scholar
- Somov G. Far-East scarlet-like fever. Moscow: Medicine; 1979.
- Laukkanen-Ninios R, Didelot X, Jolley KA, Morelli G, Sangal V, Kristo P, Population structure of the Yersinia pseudotuberculosis complex according to multilocus sequence typing. Environ Microbiol. 2011;13:3114–27. DOIPubMedGoogle Scholar
- Eitel J, Dersch P. The YadA protein of Yersinia pseudotuberculosis mediates high-efficiency uptake into human cells under environmental conditions in which invasin is repressed. Infect Immun. 2002;70:4880–91. DOIPubMedGoogle Scholar
- Isberg RR, Voorhis DL, Falkow S. Identification of invasin: a protein that allows enteric bacteria to penetrate cultured mammalian cells. Cell. 1987;50:769–78. DOIPubMedGoogle Scholar
- Hoffmann C, Pop M, Leemhuis J, Schirmer J, Aktories K, Schmidt G. The Yersinia pseudotuberculosis cytotoxic necrotizing factor (CNFY) selectively activates RhoA. J Biol Chem. 2004;279:16026–32. DOIPubMedGoogle Scholar
- Wang X, Parashar K, Sitaram A, Bliska JB. The GAP activity of type III effector YopE triggers killing of Yersinia in macrophages. PLoS Pathog. 2014;10:e1004346. DOIPubMedGoogle Scholar
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. DOIPubMedGoogle Scholar
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–2. DOIPubMedGoogle Scholar
- Portnoy DA, Martinez RJ. Role of a plasmid in the pathogenicity of Yersinia species. Curr Top Microbiol Immunol. 1985;118:29–51. DOIPubMedGoogle Scholar
- Shubin FN, Gintsburg AL, Kitaev VM, Ianishevskiĭ NV, Zenkova ZG. Analysis of the plasmid composition of Yersinia pseudotuberculosis strains and its use for typing pseudotuberculosis pathogens. Mol Gen Mikrobiol Virusol. 1989;6:20–5.PubMedGoogle Scholar
- Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci U S A. 1999;96:14043–8. DOIPubMedGoogle Scholar
- Wren BW. The Yersiniae—a model genus to study the rapid evolution of bacterial pathogens. Nat Rev Microbiol. 2003;1:55–64. DOIPubMedGoogle Scholar
1These authors were co–principal investigators.