Volume 22, Number 8—August 2016
Research
Phylogeographic Evidence for 2 Genetically Distinct Zoonotic Plasmodium knowlesi Parasites, Malaysia
Figure 3
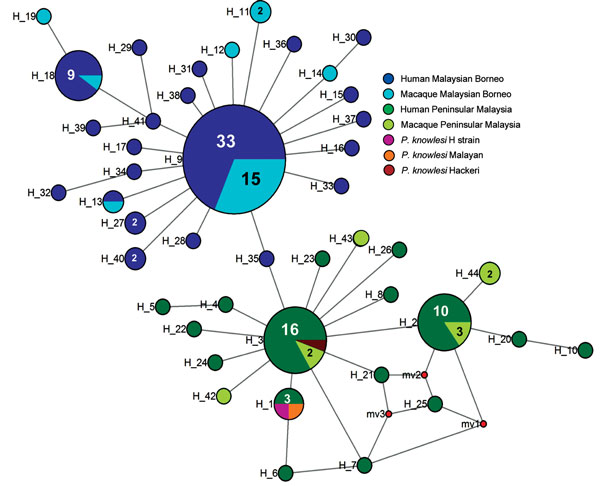
Figure 3. Median-joining networks of Plasmodium knowlesi cytochrome oxidase subunit I haplotypes from Malaysia. The genealogical haplotype network shows the relationships among the 44 haplotypes present in the 138 sequences obtained from human and macaque samples from Peninsular Malaysia and Malaysian Borneo. Each distinct haplotype has been designated a number (H_n). Circle sizes represents the frequencies of the corresponding haplotype (the number is indicated for those that were observed more than once). Small red nodes are hypothetical median vectors created by the program to connect sampled haplotypes into a parsimonious network. Distances between nodes are arbitrary.
1These authors contributed equally to this article.
Page created: July 15, 2016
Page updated: July 15, 2016
Page reviewed: July 15, 2016
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.