Volume 22, Number 8—August 2016
Dispatch
Major Persistent 5′ Terminally Deleted Coxsackievirus B3 Populations in Human Endomyocardial Tissues
Figure
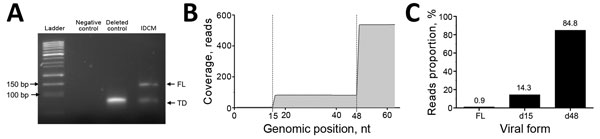
Figure. Identification of the major and minor 5′ terminally deleted or full-length enterovirus populations detected in the cardiac tissues of a patient with IDCM, Reims, France, September 2011. A) Gel electrophoresis analysis (4% agarose) of amplicons generated by using the rapid amplification of cDNA ends PCR strategy. Deleted control lane, synthetic RNA presenting with a 50-nt terminal deletion (3); IDCM lane, rapid amplification of cDNA ends PCR analysis of extracted RNA from heart tissues of the IDCM patient. B) Coverage data obtained by next-generation sequencing analysis of the cardiac tissue taken from IDCM patient. Reads obtained were filtered by bar code first to obtain 8,512,538 reads. A second discriminant step selected only reads containing both A and trP1 sequences, eliminating all artifacts resulting from early stops of the polymerase during amplification or sequencing. Following these steps, 7,354,283 reads were selected and aligned against the aligned human enterovirus group B genomes by using CLC Genomics software (CLC Bio, Aarhus, Denmark). Of these reads, only 538 were successfully aligned against enterovirus group B sequences and, more specifically, against coxsackieviruses B3 strain Nancy (Gen Bank accession no. JX312064.1). We observed that the persistent viral population in the cardiac samples consisted of a major proportion of deleted populations, with deletions ranging from 15 to 48 nt. C) Reads proportion obtained for each viral form from the IDCM patient. Coverage data were used to identify the full-length and deleted viral forms and indicate each populations’ proportions. IDCM, idiopathic dilated cardiomyopathy; FL, full-length; TD, terminally deleted.
References
- Racaniello VR. Picornaviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. Fields virology, vol. 1. 5th ed. Philadelphia: Lippincott Williams & Wilkins; 2007. p. 795–838.
- Dennert R, Crijns HJ, Heymans S. Acute viral myocarditis. Eur Heart J. 2008;29:2073–82. DOIPubMedGoogle Scholar
- Chapman NM, Kim K-S, Drescher KM, Oka K, Tracy S. 5′ terminal deletions in the genome of a coxsackievirus B2 strain occurred naturally in human heart. Virology. 2008;375:480–91. DOIPubMedGoogle Scholar
- Arbustini E, Grasso M, Porcu E, Bellini O, Diegoli M, Fasani R, Enteroviral RNA and virus-like particles in the skeletal muscle of patients with idiopathic dilated cardiomyopathy. Am J Cardiol. 1997;80:1188–93. DOIPubMedGoogle Scholar
- Kim JKS, Zhu Z, Casale G, Koutakis P, McComb RD, Swanson S, Human enterovirus in the gastrocnemius of patients with peripheral arterial disease. J Am Heart Assoc. 2013;2:e000082. DOIPubMedGoogle Scholar
- Richardson SJ, Willcox A, Bone AJ, Foulis AK, Morgan NG. The prevalence of enteroviral capsid protein VP1 immunostaining in pancreatic islets in human type 1 diabetes. Diabetologia. 2009;52:1143–51. DOIPubMedGoogle Scholar
- Lévêque N, Renois F, Talmud D, Nguyen Y, Lesaffre F, Boulagnon C, Quantitative genomic and antigenomic enterovirus RNA detection in explanted heart tissue samples from patients with end-stage idiopathic dilated cardiomyopathy. J Clin Microbiol. 2012;50:3378–80. DOIPubMedGoogle Scholar
- Cooper LT, Baughman KL, Feldman AM, Frustaci A, Jessup M, Kuhl U, The role of endomyocardial biopsy in the management of cardiovascular disease: a scientific statement from the American Heart Association, the American College of Cardiology, and the European Society of Cardiology. J Am Coll Cardiol. 2007;50:1914–31. DOIPubMedGoogle Scholar
- Nasri D, Bouslama L, Omar S, Saoudin H, Bourlet T, Aouni M, Typing of human enterovirus by partial sequencing of VP2. J Clin Microbiol. 2007;45:2370–9. DOIPubMedGoogle Scholar
- Oberste MS, Maher K, Kilpatrick DR, Pallansch MA. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol. 1999;73:1941–8.PubMedGoogle Scholar
- Sharma N, Ogram SA, Morasco BJ, Spear A, Chapman NM, Flanegan JB. Functional role of the 5′ terminal cloverleaf in Coxsackievirus RNA replication. Virology. 2009;393:238–49. DOIPubMedGoogle Scholar
- Wessely R, Klingel K, Santana LF, Dalton N, Hongo M, Jonathan Lederer W, Transgenic expression of replication-restricted enteroviral genomes in heart muscle induces defective excitation-contraction coupling and dilated cardiomyopathy. J Clin Invest. 1998;102:1444–53. DOIPubMedGoogle Scholar
- Xiong D, Yajima T, Lim B-K, Stenbit A, Dublin A, Dalton ND, Inducible cardiac-restricted expression of enteroviral protease 2A is sufficient to induce dilated cardiomyopathy. Circulation. 2007;115:94–102. DOIPubMedGoogle Scholar
- Collis PS, O’Donnell BJ, Barton DJ, Rogers JA, Flanegan JB. Replication of poliovirus RNA and subgenomic RNA transcripts in transfected cells. J Virol. 1992;66:6480–8.PubMedGoogle Scholar
- Holmblat B, Jégouic S, Muslin C, Blondel B, Joffret M-L, Delpeyroux F. Nonhomologous recombination between defective poliovirus and coxsackievirus genomes suggests a new model of genetic plasticity for picornaviruses. MBio. 2014;5:e01119–14. DOIPubMedGoogle Scholar