Volume 23, Number 5—May 2017
Research
Population Genomics of Legionella longbeachae and Hidden Complexities of Infection Source Attribution
Figure 1
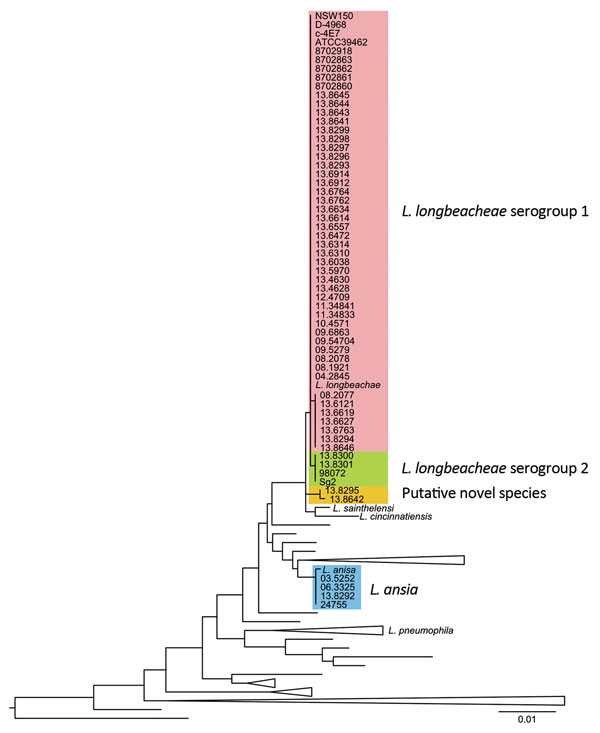
Figure 1. 6S rRNA gene–based phylogenetic tree of the sequenced genomes and all the cultured and type Legionella spp. strains available in the ribosomal database project (http://rdp.cme.msu.edu/), as accessed in May 2015. Scale bar indicates the mean number of nucleotide substitutions per site.
Page created: April 14, 2017
Page updated: April 14, 2017
Page reviewed: April 14, 2017
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.