Volume 4, Number 2—June 1998
Dispatch
Molecular and Epidemiologic Analysis of Dengue Virus Isolates from Somalia
Abstract
Nucleotide sequence analysis was performed on 14 dengue virus isolates (13 dengue-2 viruses and 1 dengue-3 virus) recovered from febrile soldiers in Somalia in 1993. The dengue-2 viruses were most closely related to dengue-2 virus recovered in Somalia in 1984. However, differences in nucleotide sequence (0.35% to 1.35%) were evident among the 1993 isolates. These differences were closely associated with the geographic location of the infection as well as with different times of infection at the same location. Genetic difference between strains was not associated with differences in clinical features. Molecular analysis of dengue viruses is a useful adjunct to epidemiologic investigation of their distribution over distance and time.
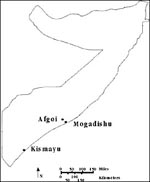
Dengue fever was a major cause of illness among U.S. troops in Somalia during 1993 (1). We identified 14 virus isolates from soldiers with dengue fever: 12 dengue-2, 1 dengue-3, and 1 mixed dengue-2 and -3 (2). Nucleotide sequence followed by phylogenetic analysis was performed to further characterize these isolates. The genotype of these virus isolates was subsequently combined with information collected during epidemiologic investigation of the febrile patients. The molecular epidemiologic analysis showed that endemic strains of dengue-2 virus were segregated in specific geographic locations (Figure 1).
Ninety patients with temperatures of at least 38.1°C who were admitted to the 86th Evacuation Army Hospital in Somalia from 27 February to 3 April 1993 underwent a standardized clinical evaluation (history and physical examination) and were asked to fill out an epidemiologic questionnaire at admission. Clinical evaluations were repeated daily until discharge. Serum samples were obtained from patients at admission and discharge and frozen at -70ºC for later testing. For this report, the geographic site of infection for each patient was defined as the site of principal activity 2 weeks before onset of illness and was assigned without knowledge of the dendrogram.
Virus isolations were performed on 81 available admission sera by injecting them into Toxorhynchites splendens mosquitoes and C6/36 cells (Aedes albopictus mosquito cell line) (3). Dengue virus isolates were identified by indirect immunofluorescence assay (IFA), by nested reverse transcriptase-polymerase chain reaction (RT-PCR) for dengue virus, and by virus neutralization (3). Virus seeds were obtained from culture fluids collected 14 days after incubation on C6/36 cells (single amplification in C6/36 cells) and stored at -70°C for genotypic analysis.
Nucleotide sequencing of the envelope (E) gene from position 639 to 1,233 was obtained for all 14 Somalia isolates. This genomic region was amplified directly by RT-PCR from extracted viral RNA and sequenced by the dye-terminator method of ABI (Applied Biosystems, Foster City, CA) (5). The nucleotide sequence was resolved by a 377 automated DNA sequencer (Applied Biosystems). The sequences, combined with the E-gene database of other viruses (5,6), were phylogenetically analyzed by genetic distance or discrete character-state methods (7). Viruses were classified into a specific serotype and genotype according to phylogenetic clustering with other dengue viruses.
The patients were men ages 19 to 39 years (median age 22); 12 were Caucasian and 2 (patients 11 and 12) were African American; all had signs and symptoms consistent with dengue fever (Table 1). The sequence analysis identified 13 dengue-2 viruses and one dengue-3 virus among the 14 isolates. These results supported previous identification by IFA and RT-PCR, except that no mixed infection with dengue-2 and -3 was found in the isolate from patient 12 after amplification in C6/36 cell culture.
The single dengue-3 isolate was recovered from a soldier stationed at the port in Mogadishu. The sequence was dengue-3 genotype III, which closely resembles Mozambique 85 strain (6) and strains from Sri Lanka, India, and Samoa.
The 13 dengue-2 isolates were all genotype IV and most closely related genetically to the 1984 isolate from Somalia (Figure 2). Phylogenetic clustering of genotype IV viruses was supported by 98% bootstrap confidence level, which also included viruses from Burkina Faso, Indonesia, Sri Lanka, and the Seychelles (5). An apparent genetic drift of 1.18% to 1.85% nucleotide sequence difference within dengue-2 viruses was observed between 1984 and 1993 (data not shown). A similar level of genetic difference (0.34% to 1.35%) was also observed among the 13 dengue-2 virus isolates collected in 1993.
Nucleotide sequence alignment from E gene position 638 to 1,233 showed 24 nucleotide changes with four amino acid alterations at the E-291, 300, 352, and 409 between 1984 and 1993 isolates (Table 2). Transition mutations were the predominant changes with a transition to transversion ratio of six to one. All 1993 isolates shared six unique nucleotide changes at position 792, 871, 885, 1,074, 1,137, and 1,161. However, only position 871, C to A change, altered amino acid sequence of Gln in the Somalia 1984 isolate to Lys in the 1993 dengue-2 virus isolates.
Viruses with certain nucleotide sequence differences appeared to be segregated by location. Virus isolates from Kismayu had a unique U change at position 1,167 that the Somalia 1984 dengue-2 virus and the Mogadishu and Afgoi isolates did not have (Table 2). Identical strains were isolated from soldiers infected in the same location at the same time. Distinguishable strains were collected from patients apparently infected at different locations within the same city, e.g., during a 1-week period at the ICRC hospital and the airport in Kismayu (Tables 1, 2). Yet, greater differences were found between dengue-2 viruses recovered from cases at different times from the same location within the city of Mogadishu. Sequences from viruses recovered in March (patient 23) or in April (patients 56, 57) were as dissimilar as sequences from viruses collected at the same time from different locations.
The data show distinct clustering of dengue-2 viruses within specific locales in Somalia. Identical dengue-2 viruses were acquired at the same time in the same locale, as shown by the four isolates from persons presumably infected at the airport in Kismayu. Moreover, virus isolates recovered from persons infected in three widely dispersed towns (Mogadishu, Kismayu, and Afgoi) were clearly distinguishable by nucleotide sequence analysis. The number of virus isolates studied in this analysis is small, and there is some imprecision in assigning the geographic location of infection. However, these data suggest a selective preference for a single circulating dengue-2 virus in a particular geographic location.
The distances between towns exceeded the flight range of mosquito vectors, unless they were transported by vehicles or by air. Movement of vectors is refuted by the consistency of isolates recovered at different locations. Additionally, no large-scale movements of troops occurred during this study. The local population was stable, and no large refugee movements occurred. Hence, the isolates reflect viruses circulating among the indigenous populations, a consequence of moving populations unexposed to dengue into endemic-disease locations.
The analysis also shows that dengue-2 viruses with unique genetic sequences may circulate at different times within the same location. The six isolates recovered from soldiers infected in Mogadishu show consistent differences over time during the short course of this analysis (2 months). These findings suggest spatial and temporal clustering of dengue viruses causing infections within Somalia. These differences are not readily explained by immigration of infective vector mosquitoes. The distances between locations within towns (e.g., 8 km) exceeded the flight range of Aedes aegypti. However, surveillance for vector mosquitoes was not routinely performed during the time of this analysis, and mosquitoes may have been carried by transport vehicles within the town.
No apparent differences in clinical features of the illness caused by these different dengue-2 strains were observed. This may reflect the small sample size, intrinsic variability in host response, and restricted time of analysis. Reports from Australia suggest that clinical features may be modified over the course of a dengue epidemic, perhaps reflecting a change in virulence of circulating virus (8). Only sporadic cases of dengue fever were detected, reflecting endemic transmission during this investigation; hence we cannot directly comment on transmission during epidemics.
Isolation of dengue viruses in C6/36 insect cells from viremic serum was required to increase virus titer for genetic analysis. Amplification of the virus isolates on insect cells may have induced or selected specific genetic changes. A comparison of the RNA T1 fingerprints of the New Guinea C isolate and C6/36 cell passage showed that under the conditions of laboratory passage in experimental hosts, during a single infection in humans, dengue-2 was subject to very little selective pressure (10). In this study, the identical isolation procedure was followed, and four identical gene sequences representing four sets of independent isolates were obtained for all isolates (Table 2); these results indicate that, if present, the same selection process was exerted on all isolation attempts.
Hence, nucleotide sequences obtained by direct sequencing of RT-PCR amplified product from serum samples of viremic patients were certainly representative of original virus genotype. Dengue-2 was the dominant virus in the mixed dengue-2, and -3 infected serum from patient 12. Failure to recover dengue-3 virus from this patient may be due to a direct growth competition of viruses in C6/36 cells. Titer of dengue-3 virus after C6/36 cell passage was insufficient to be amplified by RT-PCR directly.
The stability or relatedness of strains over time can only be surmised from these limited data. The dengue-2 viruses recovered in 1993 from this investigation are genetically distinct from the Somalia 1984 dengue-2 isolate. The extent of homology preserved for 10 years suggests that only 6 (1%) of 596 positions (Table 2) of nucleotides changed because of genetic drift. Remarkable genetic stability has been proposed for dengue viruses (10). The ability of genotyping methods to reliably resolve small (<1%) differences in sequence and the expanding access to these methods are powerful epidemiologic and clinical tools. These methods may further refine the ability to track the circulation or spread of dengue viruses within a city or country (9), as has been done with other viruses (11). These techniques may also provide insight into the evolution of dengue viruses at specific locations and perhaps detect the frequency of dual infections or the presence of recombination within dengue viruses (12).
Dr. Kanesa-thasan focuses on tropical medicine, specifically tropical virology and immunology, at the Department of Virus Diseases, Walter Reed Army Institute of Research in Washington, D.C.
References
- Sharp TW, Wallace MR, Hayes CG, Sanchez JL, DeFraites RF, Arthur RR, Dengue fever in U.S. troops during Operation Restore Hope, 19921993. Am J Trop Med Hyg. 1995;53:89–94.PubMedGoogle Scholar
- Kanesa-thasan N, Iacono-Connors L, Magill A, Smoak B, Vaughn D, Dubois D, Dengue serotypes 2 and 3 in U.S. forces in Somalia. Lancet. 1994;343:678. DOIPubMedGoogle Scholar
- Gubler DJ, Kuno G, Sather GE, Velez M, Oliver A. Mosquito cell cultures and specific monoclonal antibodies in surveillance for dengue viruses. Am J Trop Med Hyg. 1984;33:158–65.PubMedGoogle Scholar
- Tanaka M. Rapid identification of flavivirus using the polymerase chain reaction. J Virol Methods. 1993;41:311–22. DOIPubMedGoogle Scholar
- Lewis JA, Chang G-J, Lanciotti RS, Kinney RM, Mayer LW, Trent DW. Phylogenetic relationships of dengue-2 viruses. Virology. 1993;197:216–24. DOIPubMedGoogle Scholar
- Lanciotti RS, Lewis JG, Gubler DJ, Trent TW. Molecular evolution and epidemiology of dengue-3 viruses. J Gen Virol. 1994;75:65–75. DOIPubMedGoogle Scholar
- Felsenstein J. PHYLIP (Phylogeny Inference Package) [computer program] Version 3.5c. Seattle (WA): Department of Genetics, University of Washington; 1993.
- Harley D, Murray-Smith S, Weinstein P. Comparative severity of dengue symptoms in outbreaks in tropical Queensland. Commun Dis Intell. 1995;19:442–6.
- Trent DW, Grant JA, Rosen L, Monath TP. Genetic variation among dengue 2 viruses of different geographic origin. Virology. 1983;18:271–84. DOIGoogle Scholar
- Monath TP. Dengue: the risk to developed and developing countries. Proc Natl Acad Sci U S A. 1994;91:2395–400. DOIPubMedGoogle Scholar
- McDade JE, Anderson BE. Molecular epidemiology: applications of nucleic acid amplification and sequence analysis. Epidemiol Rev. 1996;18:90–7.PubMedGoogle Scholar
- Burke DS. Recombination in HIV: an important viral evolutionary strategy. Emerg Infect Dis. 1997;3:153–9. DOIGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 4, Number 2—June 1998
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Niranjan Kanesa-thasan, Department of Virus Diseases, DCD&I, Walter Reed Army Institute of Research, 14th and Dahlia Streets, N.W., Washington, DC 20307-5100, USA; fax: 202-782-0442
Top