Volume 6, Number 5—October 2000
Research
Atypical Chryseobacterium meningosepticum and Meningitis and Sepsis in Newborns and the Immunocompromised, Taiwan
Abstract
From 1996 to 1999, 17 culture-documented systemic infections due to novel, atypical strains of Chryseobacterium meningosepticum occurred in two newborns and 15 immunocompromised patients in a medical center in Taiwan. All clinical isolates, which were initially misidentified as Aeromonassalmonicida by an automated bacterial identification system, were resistant to a number of antimicrobial agents. The isolates were characterized as atypical strains of C. meningosepticum by complete biochemical investigation, 16S rRNA gene sequence analysis, cellular fatty acid analysis, and random amplified polymorphic DNA fingerprinting (RAPD). This is the first report of a cluster of atypically variant strains of C. meningosepticum, which may be an emerging pathogen in newborns and the immunocompromised.
Chryseobacterium meningosepticum, formerly known as Flavobacterium meningosepticum and CDC II-a, is a gram-negative rod widely distributed in nature. The pathogen causes meningitis in premature and newborn infants (1-3) and pneumonia, endocarditis, postoperative bacteremia, and meningitis usually associated with severe underlying illness in adults (4-7). Although C. meningosepticum infections are rare, accurate diagnosis is important because the species is usually resistant to multiple antibiotics, especially to those (including extended-spectrum -lactam agents and aminoglycosides) typically prescribed for treatment of aerobic, gram-negative bacterial infections. Moreover, epidemics may occur, and a death rate as high as 55% has been reported in a nursery outbreak (8,9).
Recent studies indicate that the species C. meningosepticum is highly heterogeneous and composed of many subgroups, which may be reclassified as separate species (10,11). Genetically defined subgroups within C. meningosepticum also differ in their pathogenicity (11). From September 1996 to March 1999, 17 culture-documented systemic infections due to a group of atypical C. meningosepticum strains occurred in two newborns and 15 immunocompromised patients in our institution. All isolates, which were initially misidentified as Aeromonas salmonicida, were resistant to a number of antimicrobial agents. The isolates were definitively identified as atypical strains of C. meningosepticum by full biochemical investigation, 16S rRNA gene sequence analysis, and cellular fatty acid analysis. Random amplified polymorphic DNA fingerprinting (RAPD) studies showed that these strains are distinct from bacteria of other genera tested, as well as from C. meningosepticum isolates from other geographic areas.
Patients and Bacterial Isolates
From September 1996 to March 1999, 17 patients admitted to Chang Gung Memorial Hospital and Children's Hospital had clinical samples positive for A. salmonicida. Primary cultures from these patients showed pure growth of nonfastidious, glucose-nonfermenting bacilli that were oxidase positive and nonmotile. The isolates were initially identified as A. salmonicida (% I.D. = 0.70) by the ID 32 GN Automatic Identification System (Biomerieux, St. Louis, MO), software version 3.2.2. Because this organism is a fish pathogen and has not been isolated in humans (12), we reviewed the charts of these patients. Clinical data for age, immunocompromising diseases, type of infection, and disease outcome were collected and analyzed. Infections were considered community acquired if the patient came to medical attention acutely ill, with an initial positive culture. Infections were considered nosocomial if cultures were negative at the time of admission or if symptomatic disease developed after the first 72 hours of hospitalization. All 17 isolates were tested biochemically and identified as C. meningosepticum with atypical reactions. These conventional biochemical tests were incubated at 30°C, and reactions were recorded after 2 days and 7 days of incubation in ambient air. Interpretation and identification were based on standard schema (13).
Clinical isolates from four index cases (strains 96, 97-1, 97-2, and 97-3) were sent to the University of Maryland and the University of Illinois at Chicago Medical Center for confirmation with conventional biochemical methods; they were again identified as C. meningosepticum.
Broth Microdilution Susceptibility Tests
Broth microdilution MIC tests were performed and interpreted by National Committee for Clinical Laboratory Standards protocols (14), including cation-adjusted Mueller-Hinton broth (Difco, Detroit, Mich.). A final inoculum of 5 ×105 CFU/mL and incubation for 16 to 20 h at 35°C in ambient air were used.
16S rRNA Gene Sequence Analysis
The MicroSeqTM 500 rDNA Bacterial Sequencing Kit (PE Biosystems, Foster City, CA) was used to amplify and sequence approximately the first 500 bp of the 16S rRNA gene, according to manufacturer's instructions. DNA sequencing with dRhodamine-labeled dye-terminators provided two overlapping strands of sequence data, with two sequencing primers. The sequence data were analyzed and assembled with AutoAssembler Software (PE Biosystems, Foster City, CA). Bacterial identification based on 16S rRNA gene sequence data, generation of the dendogram, and calculation of the percentage difference or genetic distance between sequences were performed with the MicroSeqTM Microbial Identification and Analysis Software (PE Biosystems, Foster City, CA).
Fatty Acid Analysis
Whole-cell fatty acids were extracted and analyzed (15). Analysis was performed by using an automated Hewlett-Packard HP 5890 II Microbial Identification System (MIDI, Inc., Newark, DE). Fatty acid profiles were compared with a library of cell profiles of clinically relevant bacteria making up a similarity index.
Random Amplified Polymorphic DNA Fingerprinting
This polymerase chain reaction (PCR)-based assay was performed as described (16). All bacterial strains were grown on L agar and incubated at 37°C overnight. Three loops of bacterial colony were mixed with 250 µl of Tween 20 and TE buffer and incubated at 94°C for 20 min. After addition of 250 µl of chloroform and centrifugation at 14,000 rpm for 4 min, the supernatant was collected, and the DNA was quantified by UV absorbance at A260. The primer sequence used to produce discriminatory fingerprinting profiles of bacterial strains was 5' GTCGATGTCG 3'. Each RAPD PCR reaction mixture (25 µl) contained 15 ng of genomic DNA, 40 pmol of oligonucleotides, 1 unit of Taq polymerase (GIBCO-BRL, Gaithersburg, MD), 250 µl deoxynucleoside triphosphate (Pharmacia, Laval, Quebec, Canada), 10 mM Tris-Cl (pH 8.0), 50 mM KCL, 0.001% gelatin, and 3 mM MgCl2. Each reaction mixture was overlaid with 25 µl of mineral oil and amplified with a Perkin-Elmer Cetus DNA Thermal Cycler model TC-1 according to the following profile: 4 cycles, each consisting of 5 min at 94°C, 5 min at 36°C, and 5 min at 72°C; 30 cycles, each consisting of 1 min at 94°C, 1 min at 36°C, and 2 min at 72°C; and a final extension step at 72°C for 10 min. PCR-amplified products were separated by 1.5% agarose gel electrophoresis and, after staining, were visualized and photographed under UV illumination. RAPD was performed twice on each strain, and fingerprinting was also analyzed twice. Final comparison was done by visual analysis, as well as Molecular Analyst Fingerprinting Plus Software (BIO-RAD, Ontario, Canada). We also used RAPD to examine four clinical isolates of C. meningosepticum, one Klebsiella pneumoniae, and two Burkholderia cepacia, along with the Pseudomonas aeruginosa strain P1 (16) as controls.
The first patient was a 3-day-old, full-term neonate who was transferred to our center for the treatment of early-onset neonatal sepsis and meningitis in September 1996. The infections of this index patient and of another newborn were presumed to be perinatally acquired, although culture of the cervix of the mothers was negative for this organism. Most of the other infected patients were immunocompromised (Table 1). Age >60 years and long-term hospital stay also appear to be risk factors for the infection. All infected patients <60 years of age had substantial immunosuppressive underlying diseases (Table 1). Three of the 15 nonneonatal patients died of the infection; the two newborns survived, but with severe neurologic sequelae, despite antibiotic treatment. After the first case in 1996 and three consecutive infections due to this unusual organism in early 1997, we initiated a surveillance system to monitor all infections caused by the organism in our hospital.
Biochemical properties of the isolates were nearly identical to those of typical C. meningosepticum (17), except that 8 of the 17 isolates (47%) were positive for nitrite reduction and none of them grew on MacConkey agar. Colonies of these isolates were pale yellow on blood agar plates. All the isolates were nonmotile and oxidase-positive, hydrolyzed esculin and gelatin, were positive for o-nitrophenyl-β-galactopyranoside, and produced indole. However, as described (17), the indole reaction was only weakly positive after a 48-h incubation at 30°C, and a more robust reaction was observed with inoculation into heart infusion broth rather than tryptophan broth.
The four index strains were also tested with other automated systems, including the API 20E, API 20NE, and Vitek GNI+ rapid identification systems (Biomerieux, St. Louis, MO). On API 20E, all four isolates gave a profile number of 106300400, which was interpreted as an "unacceptable profile" in the 20E database. On API 20NE, four different profile numbers were generated for the four strains (2476305, strain 96; 2476304, strain 97-1; 3476306, strain 97-2; and 2456304, strain 97-3). Profiles for three of the four strains were interpreted as "excellent identification" at 99.9% for C. meningosepticum. The profile number of 3476306 for strain 97-2 was interpreted as "doubtful" at 99.1%, but the only possible choice listed for organism identification was C. meningosepticum. By using the Vitek GNI+ card, we generated a profile number of 62022000040 for three of the four isolates, giving an interpretation of C. meningosepticum at 99%. Strain 97-2 gave a slightly different profile number of 60022000040, but with a 97% match for C. meningosepticum.
An old version of software was installed for use with the ID 32 GN Automatic Identification system (Biomerieux, St. Louis, MO) in the original identification of these isolates; this software was unable to correctly identify them as C. meningosepticum. We updated the database with a newer version 3.6.8 in May 1999. When the originally misidentified isolates were rerun through the updated database, C. meningosepticum was the first choice (%I.D. = 1.00), with A. salmonicida as the second choice (%I.D. = 0.70). No cases of A. salmonicida infection have been identified in our laboratory since May 1999.
MIC results for the 17 clinical strains from Taiwan were determined by the broth microdilution method (Table 2). All the agents tested except piperacillin and ciprofloxacin had poor in-vitro activities against these organisms. Comparative sequence analysis with the MicroSeqTM system revealed that the four index isolates share the same sequence, which clustered most closely with the type strain of C. meningosepticum (Figure 1); however, the genetic distance of 2.7% suggests that these isolates may represent either an atypical biovar of C. meningosepticum or a novel species within the genus (18). Cellular fatty acid analysis indicated that these isolates were close to C. meningosepticum, with a similarity index of 0.94. The major peaks were i-15:0 (38.5%), i-3-OH 17:0 (15.9%), and i-17:1 (7.8%).
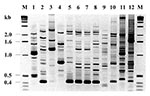
Eight RAPD fingerprints were obtained from the 17 C. meningosepticum isolates, which differed genetically, based on RAPDs, from the other four clinical C. meningosepticum strains from Canadian patients with bacteremia. The RAPD typing results of the four index isolates, 96, 97-1, 97-2, and 97-3, were compared with those of other closely related gram-negative bacteria (Figure 2). Using this typing method, we identified an apparently conserved 0.4-kb fragment in the DNA fingerprints of the 17 isolates. This genetic marker was absent from other glucose-nonfermenting bacteria, such as Pseudomonas and Burkholderia, as well as the glucose-fermenter Klebsiella, but present in one Canadian C. meningosepticum strain (Figure 2). Furthermore, C. meningosepticum, B. cepacia, P. aeruginosa strain P1, and K. pneumoniae differed in terms of their RAPD fingerprints.
Our study provides evidence of the emergence of a group of atypical strains of C. meningosepticum causing systemic infections of patients in Taiwan. This conclusion is based on phylogenetic clustering of the four index strains according to 16S rRNA gene sequence analysis and the similarity of the biochemical characteristics, cellular fatty acid profiles, and antibiograms expressed by all 17 isolates. However, unequivocal evidence of these strains as a new species within the genus Chryseobacterium will require confirmation by DNA-DNA hybridization studies.
The emergence of this specific biovar of the 17 strains has important clinical implications. First, this organism is highly pathogenic for newborn infants and immunocompromised patients, usually causing nosocomial as well as community-acquired systemic infections with substantial rates of illness and death. Second, rapid, accurate identification of the strain may be challenging for clinical microbiology laboratories. An automated bacterial identification system such as the ID 32 GN (Biomerieux, St. Louis, MO), initially misidentified all 17 isolates reported in this study as A.salmonicida. The reason for the misidentification remains unclear, although both A. salmonicida and C. meningosepticum are relatively biochemically inert and share some common biochemical characteristics, such as being esculin hydrolysis- and gelatin hydrolysis-positive and motility negative. That A. salmonicida is truly indole negative may be a clue, but a positive indole reaction for these atypical strains was difficult to obtain without special conventional test media and conditions. Because this ID 32 GN system is widely used in many clinical microbiology laboratories for the identification of gram-negative rods (19), this system should be updated or modified to improve its proficiency in identifying this as well as other emerging pathogens. Third, as with typical C. meningosepticum, choosing optimal antibiotic regimens for treating infections caused by the atypical strain was difficult because of the multidrug-resistant nature of the organism. All 17 clinical isolates were resistant to various antimicrobial agents, especially β-lactam antibiotics and aminoglycosides. The most effective drug we tested in this study was ciprofloxacin. The only other drug displaying better activity was piperacillin; however, its MIC values appeared higher, and 4 of the 17 isolates showed high-level resistance (β 16 mg/mL). The role of fluoroquinolones in the treatment of C. meningosepticum infections may be important because of their low MICs (20,21). Successful response to treatment has also been reported with trimethoprim-sulfamethoxazole, vancomycin, rifampin, clindamycin, and erythromycin (2,4,5,9), antibiotics mainly used for treating gram-positive bacterial infections. Most of these potentially effective antibiotics will not be included in the panel of susceptibility testing for any commonly isolated gram-negative bacteria other than Chryseobacterium. Therefore, accurate identification of these strains by either complete biochemical investigation or updated automated identification systems is crucial in selecting appropriate antimicrobial susceptibility testing and proper antibiotic therapy.
The 17 isolates from patients with systemic infections in Taiwan generated RAPD fingerprints that differed substantially from those of C. meningosepticum strains from Canada. Even among the 17, 8 different RAPD types were identified. Using RAPD typing, we identified an apparently conserved 0.4-kb fragment in the DNA fingerprints of the 17 isolates. This genetic marker was absent from the types of other glucose-nonfermenting bacteria, such as Pseudomonas and Burkholderia and the glucose-fermenter Klebsiella. However, one Canadian C. meningosepticum isolate had this specific genetic marker, suggesting that the atypical C. meningosepticum biovar of strains reported in this study may have been present in North America. Only a specific ribotype has been reported as an epidemiologic marker of C. meningosepticum in human infections (11). Whether the 0.4-kb band will be useful as an epidemiologic marker or represents a unique virulence factor is the subject of further studies.
In summary, we present evidence for the emergence of a cluster of atypical C. meningosepticum strains in Taiwan. This organism, which is highly pathogenic for newborns and immunocompromised patients, may be misidentified by some commercially available kit systems even at the genus level. Complete conventional biochemical testing is useful for an accurate identification. Such identification would provide clinicians with important information about the pathogenic capability of a strain and its general susceptibility profile.
Dr. Cheng-Hsun Chiu is assistant professor of Pediatrics, Chang Gung Children's Hospital and Chang Gung University. His research interests include bacterial pathogenesis and molecular epidemiology of bacterial infections.
Acknowledgments
We thank David P. Speert and Diane Roscoe for providing the Canadian clinical C. meningosepticum strains used in this study, as well as isolates of Klebsiella pneumoniae and Burkholderia cepacia.
This study was funded in part by grant NSC 89-2314-B-182A-010 from the National Science Council, The Executive Yuen, Taiwan.
References
- King EO. Studies of a group of previously unclassified bacteria associated with meningitis in infants. Am J Clin Pathol. 1959;31:241–7.PubMedGoogle Scholar
- Maderazo EG, Bassaris HP, Quintiliani R. Flavobacterium meningosepticum meningitis in a newborn infant: treatment with intraventricular erythromycin. J Pediatr. 1974;85:675–6. DOIPubMedGoogle Scholar
- Thong ML, Puthucheary SD, Lee E. Flavobacterium meningosepticum infection: an epidemiologic study in a newborn nursery. J Clin Pathol. 1981;34:429–33. DOIPubMedGoogle Scholar
- Bloch KC, Nadarajah R, Jacobs R. Chryseobacterium meningosepticum: an emerging pathogen among immunocompromised adults. Medicine. 1997;76:30–41. DOIPubMedGoogle Scholar
- Teres D. ICU-acquired pneumonia due to Flavobacterium meningosepticum. JAMA. 1974;228:732. DOIPubMedGoogle Scholar
- Olson HW, Fredricksen WC, Siboni KE. Flavobacterium meningosepticum in eight non-fatal cases of post-operative bacteremia. Lancet. 1965;1:1294–6.
- Werthamer S, Weiner M. Subacute bacterial endocarditis due to Flavobacterium meningosepticum. Am J Clin Pathol. 1971;57:410–2.PubMedGoogle Scholar
- Von Graevenitz A. Ecology, clinical significance, and antimicrobial susceptibility of infrequently encountered glucose-nonfermenting gram-negative rods. In: Gilardi GL, editor. Nonfermentative gram-negative rods: laboratory identification and clinical aspects. New York; Marcel Dekker, Inc.; 1985. p. 181-232.
- Schreckenberger PC, von Graevenitz A. Acinetobacter, Achromobacter, Alcaligenes, Moraxella, Methylobacterium, and other nonfermentative gram-negative rods. In: Murray PR, Barron EJ, Pfaller MA, Tenover FC, Yolken RH, editors. Manual of Clinical Microbiology. Seventh ed. Washington: American Society for Microbiology; 1999. p. 539-60.
- Ursing J, Bruun B. Genetic heterogeneity of Flavobacterium meningosepticum demonstrated by DNA-DNA hybridization. Acta Pathol Microbiol Immunol Scand Sect B. 1987;95:33–9.PubMedGoogle Scholar
- Colding H, Bangsborg J, Fiehn N, Bennekov T, Bruun B. Ribotyping for differentiating Flavobacterium meningosepticum isolates from clinical and environmental sources. J Clin Microbiol. 1994;32:501–5.PubMedGoogle Scholar
- Carnahan AM, Altwegg M. Taxonomy. In: Austin B, editor. The Genus Aeromonas. New York: John Wiley & Sons, Inc.; 1996. p. 1-38.
- Koneman EW, Allen DS, Janda WM, Schreckenberger PC, Winn WC Jr. Color atlas and textbook of diagnostic microbiology. 5th ed. Philadelphia: JB Lippincott Co.; 1997.
- Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. 4th ed. Approved standard M4-A4. Wayne (PA): National Committee for Clinical Laboratory Standards; 1997.
- Welch DF. Application of cellular fatty acid analysis. Clin Microbiol Rev. 1998;4:422–38.
- Mahenthiralingam E, Campbell M, Henry D, Speert DP. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J Clin Microbiol. 1996;34:129–35.PubMedGoogle Scholar
- Schreckenberger PC. Emended classification and description of the family Flavobacterium and the genus Sphingobacterium. Clin Microbiol Newsl. 1998;20:115–20. DOIGoogle Scholar
- Montgomery S, Anderson S, Waddington M, Bartell J, Nunn G, Foxall P. Variation in bacterial interspecific genetic distances--a new set of rules for interpretation of 16S rRNA sequences. In: Program and abstracts of the IXth International Congress of Bacteriological and Applied Microbiology, Sydney, Australia; 1999.
- Altwegg M, Zollinger-Iten J. Identification of Enterobacteriaceae, Aeromonas spp. and Plesiomonas shigelloides with ATB 32 GN System. J Microbiol Methods. 1987;7:103–9. DOIGoogle Scholar
- Husson MO, Izard D, Bouillet L, Lechere H. Comparative in-vitro activity of ciprofloxacin against non-fermenters. J Antimicrob Chemother. 1985;15:457–62. DOIPubMedGoogle Scholar
- Fraser S, Jorgensen JH. Reappraisal of the antimicrobial susceptibilities of Chryseobacterium and Flavobacterium species and methods for reliable susceptibility testing. Antimicrob Agents Chemother. 1997;41:2738–41.PubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 6, Number 5—October 2000
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Cheng-Hsun Chiu, Department of Pediatrics, Chang Gung Children's Hospital, 5 Fu-Shin Street, Kweishan 333, Taoyuan, Taiwan; fax: 886 3 3288957
Top