Volume 11, Number 1—January 2005
Research
Genetic Background of Escherichia coli and Extended-spectrum β-Lactamase Type
Figure
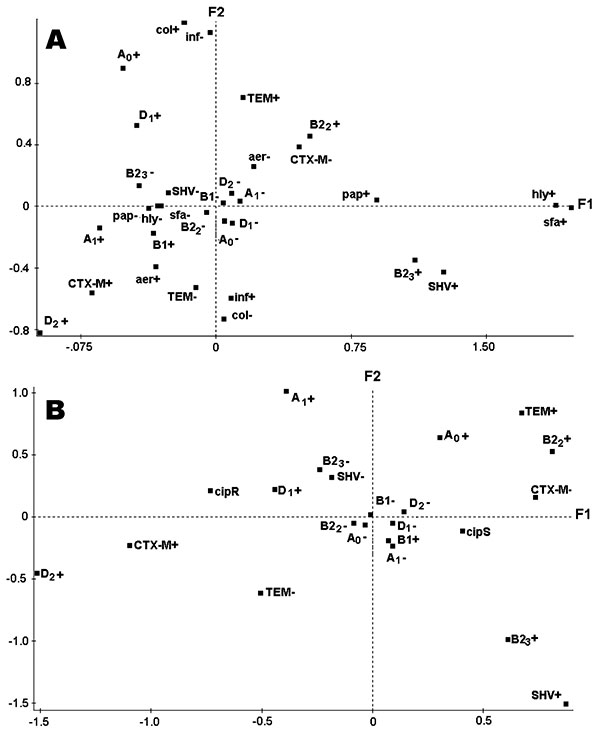
Figure. Graphic representation of the results of the factorial analysis of correspondence carried out with whole data from the 129 Escherichia coli strains. A) Projections of the variables on the F1/F2 plane: phylogenetic group and subgroups (A0, A1, B1, B22, B23, D1, and D2), type of extended-spectrum β-lactamase (ESBL) (TEM, SHV, CTX-M), virulence factors (pap, sfa, hly, aer), and the source infection (inf) or colonization (col). B) Projections of the variables on the F1/F2 plane: phylogenetic group and subgroups (A0, A1, B1, B22, B23, D1, and D2), type of ESBL (TEM, SHV, CTX-M), and ciprofloxacin resistance (cipR) or the ciprofloxacin susceptibility (cipS).
Page created: April 14, 2011
Page updated: April 14, 2011
Page reviewed: April 14, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.