Volume 15, Number 12—December 2009
Synopsis
Towards Control of Streptococcus iniae
Abstract
Streptococcus iniae is an emerging zoonotic pathogen; such infections generally occur through injuries associated with preparing whole fresh fish for cooking. Those infected to date have been of Asian descent, are usually elderly (average age 68 years), and have had >1 underlying conditions that may predispose them to infection. Studies of the foundations of growth characteristics of S. iniae and its interactions with piscine host cells have recently been complemented by molecular studies. Advances in molecular biology have allowed research groups to identify numerous virulence factors and to explore their roles in the progression of S. iniae infection. Many of these virulence factors are homologous to those found in the major human pathogen S. pyogenes. An increased understanding of the properties of these factors and their effect on the success of infection is leading to novel approaches to control S. iniae infection; in particular, vaccination programs at fish farms have reduced the reservoir of infection for additional clinical cases.
Streptococcus iniae is a major fish pathogen in many regions of the world. These bacteria are also zoonotic with infections in humans associated with the handling and preparation of infected fish. The first human infections were reported in 1996 (1), and S. iniae was noted as an emerging zoonotic disease transmitted by food animals at the International Conference on Emerging Infectious Diseases in 2000 (2). Human infections with S. iniae have been sporadic but continue to be reported with new cases arising in 2009 (3). Reports of these cases are likely to increase because of enhanced awareness, more reliable detection and identification methods, and the global expansion of finfish aquaculture. Most cases of human S. iniae infections have been in persons of Asian descent, who are elderly and commonly have >1 underlying conditions such as diabetes mellitus, chronic rheumatic heart disease, cirrhosis, or other conditions (1,3–7).
Carrier fish have been implicated in fish-to-fish transmission of S. iniae (8), and these carriers may be responsible for human infection because fish with overt signs of disease are unmarketable. Soft tissue injuries that occur during the preparation of fresh fish from wet markets usually result in bacteremic cellulitis of the hand, followed by >1 of these conditions: endocarditis, meningitis, arthritis, sepsis, pneumonia, osteomyelitis, and toxic shock (7). Infections are treated with a course of antimicrobial drugs such as penicillin, ampicillin, amoxicillin, cloxacillin, cefazolin, and/or gentamicin, doxycycline, and trimethoprim/sulfamethoxazole over a period of 1 to several weeks, depending on the nature of the infection (3–5,9). S. iniae is not currently assigned to any Lancefield group and is β-hemolytic on blood agar, with some clinical strains isolated from Asia being more mucoid than others (6).
Underreporting of human cases is likely because identification of S. iniae is based on biochemical testing of isolates with commercial kits; the use of kits is associated with problems because S. iniae is not listed in commercial or clinical databases, and many atypical strains are assigned low matches (1,4). According to the Australian Institute of Health and Welfare (www.aihw.gov.au), between 1999–2000 and 2006–2007, a total of 2,824 cases of “other” or “unspecified” streptococcal sepsis required hospitalization in Australia that were attributed to nongroup A, B, or C streptococci, or S. pneumoniae, and 2,026 cases were in persons >50 years of age. During the same period, the trend in the number of cases per year attributable to other or unspecified streptococci has been upward, rising from a total of 278 cases in 1999–2000 to 430 in 2006–2007 (155% increase). In the >50 years age group, this upward trend is more pronounced, with a 168% increase in cases requiring hospitalization. It is therefore probable that some cases of S. iniae infection in Australia in the at-risk age group have been misidentified. Misidentification of S. iniae infection is likely to be the main reason for low levels of detection because most cases of this emerging pathogen are detected during retrospective studies specifically targeting S. iniae. This finding is likely to be the case in countries around the world that have reported outbreaks in fish farms, but no human cases to date. Molecular-based detection and identification methods have recently been developed (information on this aspect of identification can be found in a recent review by Agnew and Barnes) and these will lead to improved reporting in future years (10).
Observations on the epidemiology and pathogenesis of S. iniae infections are still ongoing; however, valuable information on the differentiation of strains (as being either commensal or pathogenic) has benefitted research. Because of the lack of potential virulence factors or phenotypic differences between commensal and pathogenic strains, pulsed-field gel electrophoresis (PFGE) analysis showed that differences existed between human clinical isolates and those from fish surfaces (1). The human clinical isolates showed little variation between one another, while considerable differences were found between the 2 American Type Culture Collection (Manassas, VA, USA) dolphin strains and 32 other fish isolates. It was determined that some unknown factors important to pathogenicity were not present in all strains (1). Little variation has also been found between clinical strains from the United States and Canada, although 1 strain from Pennsylvania had a PFGE pattern similar to the type strain (11), and 2 clinical isolates from Hong Kong were unrelated to a strain from Canada (4). In a PFGE study of isolates from Australia, similarities between fish pathogenic isolates and human clinical isolates from North America, in addition to multiple genotypes between and within different fish farms, were reported (12). Another PFGE study of strains from a variety of species of diseased fish in the People’s Republic of China found that there were 17 genotypes from 27 strains clustering into 5 major groups (13). As with the study from Australia, multiple genotypes were found between and within different farms.
Clinical isolates from the United States were able to multiply by 2 to 5 generations in 3 hours in fresh human blood; however, 2 isolates from Canada were able to survive, but not multiply, in human blood (11). Resistance to phagocytic killing in whole blood by pathogenic strains of S. iniae contrasted with isolates identified as commensal strains that were susceptible (9). The pathogenic strain 9117 (a human clinical isolate causing cellulitis) caused weight loss in experimentally infected mice and was highly cellulytic to human brain microvessel endothelial cell (BMEC) monolayers and invasive of Hep-2 cells. However, adherence to, and invasion of, BMEC cells by strain 9117 was lower than that for commensal strain 9066 (obtained in a swab sample from a healthy fish) (9). In a similar study, pathogenic strains were more resistant to oxidative burst activity in macrophages (14).
The mode of invasion of S. iniae was studied in skin epithelial cell monolayers of rainbow trout viewed under polarized light (15). S. iniae adhered to, then invaded epithelial cells, but its persistence and replication inside the cells was short-lived. However, transcytosis from epithelial cells occurred within 30 minutes of contact without damaging the cells or cellular junctions (15). Once the epithelial cell layer had been breached, dissemination throughout the fish through internalization in macrophages can occur. By inducing macrophage death, this process is one of the most efficient ways of transporting the infection into the brain (15,16).
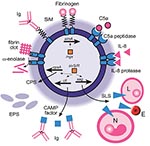
Recent molecular research into factors that contribute to the virulence of S. iniae has identified several candidates, including surface proteins, capsular polysaccharides, and extracellular secreted products (Figure). Moreover, the recent sequencing of the complete genome of S. iniae will accelerate the discovery of additional virulence factors and lead to identification of targets for effective vaccines for farmed fish, thus reducing the potential for zoonotic infection. In light of the recent rapid increase in our knowledge of this emerging pathogen, we will present a synopsis of the processes involved in infection that have been elucidated to date.
SiM Protein
M proteins are one of the major virulence factors in group A streptococci (S. pyogenes; GAS). The high level of diversity of emm gene types (a hypervariable gene encoding the M protein) has contributed to the success of GAS in causing infections in humans. The M-like protein from S. iniae, SiM, is also a prime candidate in virulence (17,18). The SiM protein is a coiled-coil protein that has a molecular mass of ≈53 kDa, although 2 other variants, one with a 1-aa insertion between the coils and another with a much larger mass of ≈59 kDa, have also been described (17). An additional variant had a natural frameshift/premature termination of the SiMA1 type in a strain from an infected tilapia (18). All SiM proteins possess the classical gram-positive membrane anchor motif LPXTG, and although they have several repeat motifs in the coil regions, they are not in tandem as in M proteins from other species. In common with M proteins from S. pyogenes, S. equi, and S. dysgalactiae, SiM protein is a surface protein that binds human fibrinogen to protect the bacterium from phagocytic activity (17). SiM proteins may also bind trout immunoglobulin by the Fc region (19). An allelic exchange study showed that SiM protein is a major virulence factor of S. iniae, contributes to adherence to fish epithelial cells (18), and also confirmed earlier observations that SiM contributes to macrophage resistance (17).
The SiM protein genes, simA and simB, are likely to be regulated by a multigene regulatory protein, Mgx, that is homologous to the Mga protein found in GAS (17). There are sequence elements upstream of the simA and simB genes that are similar to DNA binding sites described for Mga. The finding of a second mga-like element (mgx2) immediately downstream of mgx in a tilapia brain abscess isolate may represent part of an alternative virulence strategy (18).
C5a Peptidase
C5a peptidase hydrolyses the neutrophil chemoattractant complement factor C5a (18) and thus impairs the ability of the infected host to fight the infection. C5a peptidase is a surface protein with a LPXTN gram-positive anchor motif (18). In GAS, C5a peptidase is found in culture supernatants; however, this observation has not been made for S. iniae (18). In S. iniae, C5a peptidase is a 123-kDa protein, encoded by scpI, with similar structural features and conserved residue positions to the GAS counterpart (18). C5a peptidase may have arisen in S. iniae by horizontal gene transfer, given its close proximity to a transposase and similar genetic organization found in GAS (18). Notably, allelic exchange has shown that this protein by itself is not required for virulence in fish and its role in pathogenesis is likely a minor one (18).
Interleukin-8 Protease
Interleukin-8 ( IL-8) is produced in the host in response to stimuli such as lipopolysaccharides, viruses, and other cytokines. IL-8 protease is a cell envelope protease that is able to degrade the chemokine IL-8 and results in increased neutrophil resistance and disease dissemination (20). It is encoded by the cepI gene, resulting in a 1,631-aa protein with a C-terminal LPXTG gram-positive anchor motif and is homologous to the cepA gene in GAS (20).
Streptolysin S
The ability of S. iniae to hemolyse erythrocytes and damage host cell membranes results from the activity of cytolysins (21). The cytolysin possessed by S. iniae is homologous to streptolysin S (SLS) from GAS (21) and affects erythrocytes, neutrophils, lymphocytes, and some tissue types in tissue culture (22), but does not have roles in phagocytic resistance nor epithelial cell adherence and invasion (23). Nine genes in the sag operon are involved in SLS formation; these share 73% homology with GAS SLS genes (21). The number of genes and their genetic order are identical in both microorganisms (21). The sagA gene encodes a peptide that is 73% identical to the sagA protein from GAS, and the sagB gene encodes a protein with 77% identity to the sagB protein from GAS, which is predicted to use flavin mononucleotide as a cofactor (21). The sagC-F genes are similar to their counterpart genes in GAS, and the sagG-I genes encode ATP binding cassette-type (ABC) transport systems (21). Other sequence features, such as inverted repeats between the sagA and sagB genes and after the sagI gene, have similarities with GAS sag operon genes (21). All genes in the operon are required to produce SLS as knockout of sagB in S. iniae caused loss of hemolytic activity (21). Likewise, when the S. iniae sagA gene was transformed into a nonhemolytic allelic mutant strain of GAS (NZ131 sagA∆cat), the S. iniae version of the sagA gene restored hemolytic activity (21,23). The cytotoxic properties of SLS toward fish cells and the likely promotion of cerebrovascular trauma represent a major virulence factor in the pathogenesis of S. iniae (23).
The sagA gene in S. iniae is regulated by a 2-component signal transduction system called sivS/R (24). sivS/R regulates virulence in vivo because no deaths occurred in a mouse infection model when mice were infected with a deletion mutant of pathogenic strain 9117 (9117∆siv) compared with 75% deaths when mice were infected with the wild type strain (24). In the siv deletion mutant, sagA expression was decreased by 3-fold. sivS/R also regulates the expression of surface proteins, including a lipoprotein/ABC transporter homologous to Spy1228, pyruvate kinase, and a hyaluronate-associated protein homologous to that from S. equi or an ABC transporter from S. agalactiae (24). However, the role for these proteins in a virulence setting is hitherto unknown in S. iniae.
CAMP Factor
The pore-forming toxin CAMP factor synergistically acts with sphingomylinase-producing Staphylococcus aureus to produce a distinct arrow-shaped area of complete lysis of erythrocytes on sheep blood agar (24). CAMP factor has also been shown to bind immunoglobulin by the Fc region and therefore contributes to virulence (24). S. iniae harbors a CAMP factor–like gene, cfi, (24) that encodes a peptide of ≈27 kDa and shares 62% identity with cfa from GAS that is regulated by the sivS/R system. Knockout mutants of the sivS/R system resulted in a reduced lytic reaction, and real-time PCR analysis of cfi gene expression showed it was expressed at only 10% of wild-type levels (24).
Immunoglobulin-Binding Proteins
A putative protein G-like protein, a cell wall associated protein first identified from group G streptococci (≈70 kDa), from S. iniae was capable of binding trout immunoglobulin only when grown in the presence of trout serum (19). Proteins of ≈35 kDa, ≈70 kDa, and >100 kDa were found to bind trout immunoglobulin. The size of one of the detected bands >100 kDa is similar in size to the tetramer formation of SiM proteins (17); however, experimental evidence is needed to confirm this. CAMP factor is also known to bind immunoglobulin (24).
Capsule
One of the most effective ways for a bacterium to avoid phagocytosis is by the production of capsular polysaccharide (CPS), and strains with CPS are more virulent than their unencapsulated counterparts (25–27). The presence of capsule is also involved in inhibiting complement C3 deposition (27). Miller and Neely (27) used signature tagged mutagenesis to identify virulence genes using a zebrafish (Danio rerio) model. Five attenuated mutants with unique insertions in polysaccharide synthesis genes with homology to those found in S. thermophilus plus 2 additional clones with insertions in a homologous gene near capsule synthesis genes from Bacteroides thetaiotaomicron were found. In contrast to the wild-type strain 9117, these mutants aggregated in broth culture with chain lengths up to 4× longer than the wild type. Differences in buoyancy due to degree of encapsulation showed that the production of excess capsule is likely to be as detrimental to survival as too little capsule (27).
The capsule operon of S. iniae is ≈21 kb in size and consists of 21 open reading frames (28). The genes have homology to genes found in other streptococci such as S. pyogenes, S. agalactiae, S. suis, and S. thermophilus (28). An insertion sequence, IS981, was found between cpsL and cpsM in strain 9117 (28). cpsY (78% identity to CpsY in GAS) precedes cpsA and is transcribed in the opposite direction and is thought to be the promoter of the capsule operon (as well as other virulence genes) because it has a high level of homology to the LysR transcriptional regulators (28). A major difference was found in the operons from the virulent strain 9117 and commensal strain 9066 with strain 9066 having an ≈10-kb deletion missing the genes cpsF-L and orf276, orf193, and orf151. In addition, the cpsM gene in the commensal strain 9066 was truncated with the first 154 nt being absent, which casts doubt on the functionality of the gene (28).
The genes cpsA-E are responsible for the length of the monosaccharide sugar chains and their export (29,30). The central region of the operon from cpsF to cpsL contains several genes encoding glycosyl transferases, which have a role in the polymerization of the capsule chain (28). This region is where most mutations in the virulent strain 9117 were found to play a role in pathogenesis (27) and is the same region where the deleted genes in the commensal strain 9066 occurred (28). The G + C content of the operon genes varies widely from 22% to 40% (28). The cpsF-L region has a G + C content of ≈27%, which is lower than that found for other S. iniae sequences and may indicate horizontal acquisition from other members of the Firmicutes (28).
Mutations in cpsA in strain 9117 resulted in long chain formation, aggregation of cells in broth culture and high buoyancy characteristic of reduced encapsulation (28). The cpsA mutant was attenuated in both brain and heart tissues. A cpsY mutant was slightly less encapsulated than the wild type and was attenuated only in heart tissue (28).
Allelic exchange of the cpsD gene resulted in reduced capsule, increased chain length, a marked decrease in all capsular monosaccharides, and a high degree of attenuation (31). cpsD encodes an autophosphorylating tyrosine kinase thought to be responsible for capsule polymerization and export (31). cpsD mutants were able to bind more effectively to host tissues, such as epithelial cells, due to loss of overall negative charge (31). Insertions in the cpsH and cpsM genes have resulted in underproduction of capsule and overproduction of capsule, respectively (28). In a flounder isolate obtained in Japan, insertions in cpsH, cpsM, cpsI, and orf276 of strain NUF631 resulted in attenuation, which was measured by increased chemiluminescence response of macrophages and loss of acidic polysaccharides (26). Like CAMP factor and streptolysin S, the 2-component system sivS/R is involved in transcriptional regulation of the capsular operon (32).
Phosphoglucomutase
Also involved in capsular biosynthesis is phosphoglucomutase, a 571-aa protein encoded by pgmA that interconverts glucose 6-phosphate and glucose 1-phosphate (33). Transposon mutagenesis of the promoter region upstream of pgmA resulted in a highly attenuated mutant that was more susceptible to whole blood killing (33). This susceptibility was attributed to a decrease in the amount of exopolysaccharide capsule on the cell surface, decreased negative charge, and a larger cell volume 3–5× that of the wild type. Increased susceptibility to the pore-forming cationic antimicrobial peptide moronecidin was also reported, most likely due to changes in cell wall architecture because of increased cellular volume and a decrease in cell wall rigidity (33).
Exopolysaccharide
The quantitative composition of monosaccharides present in exopolysaccharide (EPS) is distinct from those found in CPS (34). The routine vaccination of fish in Israel has given rise to new strains of S. iniae responsible for mass fish deaths (34). These new strains were formed when an autogenous vaccine strain, KFP404, was succeeded by new strains KFP468, KFP477, and KFP523, which were characterized by a viscous broth culture similar to that observed with S. thermophilus used in yoghurt production (34). EPS production by the successor strains was 5× higher than the autogenous vaccine strain. Vaccination of fish with the EPS extracts elicited a survival rate of 78%, which was similar to the 72% survival rate when whole cells were used. Thus, EPS appears to be antigenic and excessive production may have been selected by vaccination (34).
α-Enolase
The ability of S. iniae to cross tissues through plasminogen activation is facilitated by α-enolase (35), which is also a known contributory factor to the virulence of GAS (36). The proteolytic activity of plasmin in dissolving fibrin clots enables pathogens to migrate faster through extracellular matrices (37), and α-enolase expedites invasion through the host tissues (38) and, ultimately, into the circulatory system. The α-enolase from S. iniae (≈50 kDa) is a plasmin/plasminogen binding enzyme that is 97% similar to the α-enolases of S. agalactiae and GAS (35). Immunoblot using antibodies raised against the purified recombinant protein showed cell wall association in S. iniae; however, it does not contain the classical gram-positive membrane anchor.
S. iniae opportunistically infects elderly persons with serious underlying conditions. The expression of a suite of virulence factors, many of them similar to those found in GAS, is responsible for successful entry, propagation, and evasion of immune defenses of the host by this bacterium. Another virulence factor, polysaccharide deacetylase, encoded by the pdi gene, has been recently described (39). With the global rise of aquaculture and the dependence on it to provide food in many areas of the world, the numbers of cases of S. iniae infection are likely to be much higher than currently reported and will increase in the future with the expansion of the industry. Understanding the pathogenic processes of S. iniae is already facilitating the development of vaccines for use in fish farms and represents the most sustainable and effective method of reducing the incidence of economically devastating outbreaks and clinical presentations in humans, especially in those most at risk.
Dr Baiano is a research officer in the Centre for Marine Studies at The University of Queensland. He is a microbial ecologist and has research interests in marine microbiology, aquatic animal health, and aquaculture.
Dr Barnes is associate professor in aquatic animal health at The University of Queensland, Centre for Marine Studies and School of Biological Sciences. His research interests include marine microbiology, comparative immunology, and host–pathogen interactions in marine animals.
References
- Weinstein MR, Litt M, Kertesz DA, Wyper P, Rose D, Coulter M, Invasive infections due to a fish pathogen, Streptococcus iniae. S. iniae study group. N Engl J Med. 1997;337:589–94. DOIPubMedGoogle Scholar
- Hansen GR, Woodall J, Brown C, Jaax N, McNamara T, Ruiz A. Emerging zoonotic diseases. Panel summary from the International Conference on Emerging Infectious Diseases Conference, Atlanta, Georgia, 2000. Emerg Infect Dis. 2001;7:537. DOIPubMedGoogle Scholar
- Koh TH, Sng LH, Yuen SM, Thomas CK, Tan PL, Tan SH, Streptococcal cellulitis following preparation of fresh raw seafood. Zoonoses Public Health. 2009;56:206–8. DOIPubMedGoogle Scholar
- Lau SKP, Woo PCY, Tse H, Leung K-W, Wong SSY, Yuen K-Y. Invasive Streptococcus iniae infections outside North America. J Clin Microbiol. 2003;41:1004–9. DOIPubMedGoogle Scholar
- Koh TH, Kurup A, Chen J. Streptococcus iniae discitis in Singapore. Emerg Infect Dis. 2004;10:1694–5.PubMedGoogle Scholar
- Lau SKP, Woo PCY, Luk W-K, Fung AMY, Hui W-T, Fong AHC, Clinical isolates of Streptococcus iniae from Asia are more mucoid and beta-hemolytic than those from North America. Diagn Microbiol Infect Dis. 2006;54:177–81. DOIPubMedGoogle Scholar
- Sun JR, Yan JC, Yeh CY, Lee SY, Lu JJ. Invasive infection with Streptococcus iniae in Taiwan. J Med Microbiol. 2007;56:1246–9. DOIPubMedGoogle Scholar
- Zlotkin A, Hershko H, Eldar A. Possible transmission of Streptococcus iniae from wild fish to cultured marine fish. Appl Environ Microbiol. 1998;64:4065–7.PubMedGoogle Scholar
- Fuller JD, Bast DJ, Nizet V, Low DE, de Azavedo JC. Streptococcus iniae virulence is associated with a distinct genetic profile. Infect Immun. 2001;69:1994–2000. DOIPubMedGoogle Scholar
- Agnew W, Barnes AC. Streptococcus iniae: an aquatic pathogen of global veterinary significance and a challenging candidate for reliable vaccination. Vet Microbiol. 2007;122:1–15. DOIPubMedGoogle Scholar
- Facklam R, Elliot J, Shewmaker L, Reingold A. Identification and characterization of sporadic isolates of Streptococcus iniae isolated from humans. J Clin Microbiol. 2005;43:933–7. DOIPubMedGoogle Scholar
- Nawawi RA, Baiano J, Barnes AC. Genetic variability amongst Streptococcus iniae isolates from Australia. J Fish Dis. 2008;31:305–9. DOIPubMedGoogle Scholar
- Zhou SM, Xie MQ, Zhu XQ, Ma Y, Tan ZL, Li AX. Identification and genetic characterization of Streptococcus iniae strains isolated from diseased fish in China. J Fish Dis. 2008;31:869–75. DOIPubMedGoogle Scholar
- Buchanan JT, Colvin KM, Vicknair MR, Patel SK, Timmer AM, Nizet V. Strain-associated virulence factors of Streptococcus iniae in hybrid-striped bass. Vet Microbiol. 2008;131:145–53. DOIPubMedGoogle Scholar
- Eyngor M, Chilmonczyk S, Zlotkin A, Manuali E, Lahav D, Ghittino C, Transcytosis of Streptococcus iniae through skin epithelial barriers: an in vitro study. FEMS Microbiol Lett. 2007;277:238–48. DOIPubMedGoogle Scholar
- Zlotkin A, Chilmonczyk S, Eyngor M, Hurvitz A, Ghittino C, Eldar A. Trojan horse effect: phagocyte-mediated Streptococcus iniae infection of fish. Infect Immun. 2003;71:2318–25. DOIPubMedGoogle Scholar
- Baiano JCF, Tumbol RA, Umapathy A, Barnes AC. Identification and molecular characterisation of a fibrinogen binding protein from Streptococcus iniae. BMC Microbiol. 2008;8:67. DOIPubMedGoogle Scholar
- Locke JB, Aziz RK, Vicknair MR, Nizet V, Buchanan JT. Streptococcus iniae M-like protein contributes to virulence in fish and is a target for live attenuated vaccine development. PLoS One. 2008;3:e2824. DOIPubMedGoogle Scholar
- Barnes AC, Horne MT, Ellis AE. Streptococcus iniae expresses a cell surface non-immune trout immunoglobulin-binding factor when grown in normal trout serum. Fish Shellfish Immunol. 2003;15:425–31. DOIPubMedGoogle Scholar
- Zinkernagel AS, Timmer AM, Pence MA, Locke JB, Buchanan JT, Turner CE, The IL-8 protease SpyCEP/ScpC of group A streptococcus promotes resistance to neutrophil killing. Cell Host Microbe. 2008;4:170–8. DOIPubMedGoogle Scholar
- Fuller JD, Camus AC, Duncan CL, Nizet V, Bast DJ, Thune RL, Identification of a streptolysin S–associated gene cluster and its role in the pathogenesis of Streptococcus iniae disease. Infect Immun. 2002;70:5730–9. DOIPubMedGoogle Scholar
- Nizet V, Beall B, Bast DJ, Datta V, Kilburn L, Low DE, Genetic locus for streptolysin S production by group A Streptococcus. Infect Immun. 2000;68:4245–54. DOIPubMedGoogle Scholar
- Locke JB, Colvin KM, Varki N, Vicknair MR, Nizet V, Buchanan JT. Streptococcus iniae β-hemolysin streptolysin S is a virulence factor in fish infection. Dis Aquat Organ. 2007;76:17–26. DOIPubMedGoogle Scholar
- Bolotin S, Fuller JD, Bast DJ, de Azavedo JCS. The two-component system sivS/R regulates virulence in Streptococcus iniae. FEMS Immunol Med Microbiol. 2007;51:547–54. DOIPubMedGoogle Scholar
- Kanai K, Notohara M, Kato T, Shutou K, Yoshikoshi K. Serological characterization of Streptococcus iniae strains isolated from cultured fish in Japan. Fish Pathology. 2006;41:57–66. DOIGoogle Scholar
- Shutou K, Kanai K, Yoshikoshi K. Virulence attenuation of capsular polysaccharide-deleted mutants of Streptococcus iniae in Japanese flounder Paralicthys olivaceus. Fish Pathology. 2007;42:42–8.
- Miller JD, Neely MN. Large-scale screen highlights the importance of capsule for virulence in the zoonotic pathogen Streptococcus iniae. Infect Immun. 2005;73:921–34. DOIPubMedGoogle Scholar
- Lowe BA, Miller JD, Neely MN. Analysis of the polysaccharide capsule of the systemic pathogen Streptococcus iniae and its implications in virulence. Infect Immun. 2007;75:1255–64. DOIPubMedGoogle Scholar
- Griffin AM, Morris VJ, Gasson MJ. The cpsABCDE genes involved in polysaccharide production in Streptococcus salivarius ssp. thermophilus strain NCBF2393. Gene. 1996;183:23–7. DOIPubMedGoogle Scholar
- Morona JK, Miller DC, Morona R, Paton JC. The effect that mutations in the conserved capsular regions polysaccharide biosynthesis genes cpsA, cpsB, and cpsD have on virulence of Streptococcus pneumoniae. J Infect Dis. 2004;189:1905–13. DOIPubMedGoogle Scholar
- Locke JB, Colvin KM, Datta AK, Patel SK, Naidu NN, Neely MN, Streptococcus iniae capsule impairs phagocytic clearance and contributes to virulence in fish. J Bacteriol. 2007;189:1279–87. DOIPubMedGoogle Scholar
- Bolotin S, Fuller JD, Bast DJ, Beveridge TJ, de Azavedo JCS. Capsule expression regulated by a two-component signal transduction system in Streptococcus iniae. FEMS Immunol Med Microbiol. 2007;50:366–74. DOIPubMedGoogle Scholar
- Buchanan JT, Stannard JA, Lauth X, Ostland VE, Powell HC, Westerman ME, Streptococcus iniae phosphoglucomutase is a virulence factor and a target for vaccine development. Infect Immun. 2005;73:6935–44. DOIPubMedGoogle Scholar
- Eyngor M, Tekoah Y, Shapira R, Hurvitz A, Zlotkin A, Lublin A, Emergence of novel Streptococcus iniae exopolysaccharide-producing strains following vaccination with nonproducing strains. Appl Environ Microbiol. 2008;74:6892–7. DOIPubMedGoogle Scholar
- Kim MS, Choi SH, Lee EH, Nam YK, Kim SK, Kim KH. α-enolase, a plasmin(ogen) binding protein and cell wall associating protein from a fish pathogenic Streptococcus iniae strain. Aquaculture. 2007;265:55–60. DOIGoogle Scholar
- Pancholi V, Fischetti VA. Alpha-enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J Biol Chem. 1998;273:14503–15. DOIPubMedGoogle Scholar
- Eberhard T, Kronvall G, Ullberg MBB. Surface bound plasmin promotes migration of Streptococcus pneumoniae through reconstituted basement membranes. Microb Pathog. 1999;26:175–81. DOIPubMedGoogle Scholar
- Lottenberg R. A novel approach to explore the role of plasminogen in bacterial pathogenesis. Trends Microbiol. 1997;5:466–7. DOIPubMedGoogle Scholar
- Milani CJ, Aziz RK, Locke JB, Dahesh S, Nizet V, Buchanan JT. The novel polysaccharide deacetylase homolog Pdi contributes to virulence of the aquatic pathogen Streptococcus iniae. Microbiology. 2009. In press .DOIPubMedGoogle Scholar
Figure
Cite This ArticleTable of Contents – Volume 15, Number 12—December 2009
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Justice C.F. Baiano, The University of Queensland, Aquatic Animal Health Laboratory, Centre for Marine Studies, St. Lucia, Queensland 4072, Australia
Top