Volume 17, Number 12—December 2011
Research
Novel Multiplexed HIV/Simian Immunodeficiency Virus Antibody Detection Assay
Figure 2
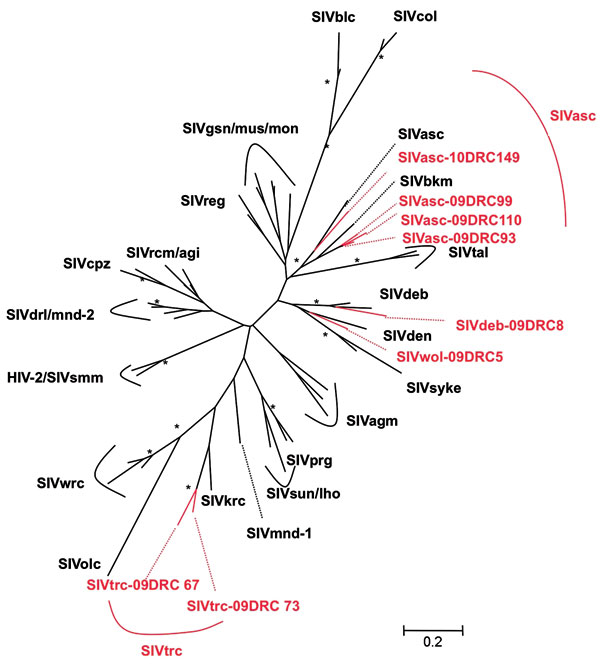
Figure 2. Phylogenetic relationships of the newly derived simian immunodeficiency virus (SIV) sequences in pol to representatives of the other SIV lineages. Newly identified strains in this study are in red and reference strains are in black. Unrooted trees were inferred from 350-bp nucleotides. Analyses were performed by using discrete gamma distribution and a generalized time reversible model. The starting tree was obtained by using phyML (27). One hundred bootstrap replications were performed to assess confidence in topology. Numbers at nodes are from 100 bootstrap replicates; only those >90% are shown with an asterisk. Scale bar represents nucleotide replacements per site.
References
- Hahn BH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–14. DOIPubMedGoogle Scholar
- Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML, Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313:523–6. DOIPubMedGoogle Scholar
- Van Heuverswyn F, Li Y, Neel C, Bailes E, Keele BF, Liu W, Human immunodeficiency viruses: SIV infection in wild gorillas. Nature. 2006;444:164. DOIPubMedGoogle Scholar
- Gao F, Yue L, White AT, Pappas PG, Barchue J, Hanson AP, Human infection by genetically diverse SIVSM-related HIV-2 in west Africa. Nature. 1992;358:495–9. DOIPubMedGoogle Scholar
- Hirsch VM, Olmsted RA, Murphey-Corb M, Purcell RH, Johnson PR. An African primate lentivirus (SIVsm) closely related to HIV-2. Nature. 1989;339:389–92. DOIPubMedGoogle Scholar
- Plantier JC, Leoz M, Dickerson JE, De Oliveira F, Cordonnier F, Lemée V, A new human immunodeficiency virus derived from gorillas. Nat Med. 2009;15:871–2. DOIPubMedGoogle Scholar
- Wolfe ND, Prosser TA, Carr JK, Tamoufe U, Mpoudi-Ngole E, Torimiro JN, Exposure to nonhuman primates in rural Cameroon. Emerg Infect Dis. 2004;10:2094–9.PubMedGoogle Scholar
- Peeters M, Courgnaud V, Abela B, Auzel P, Pourrut X, Bibollet-Ruche F, Risk to human health from a plethora of simian immunodeficiency viruses in primate bushmeat. Emerg Infect Dis. 2002;8:451–7.PubMedGoogle Scholar
- Calattini S, Betsem EB, Froment A, Mauclère P, Tortevoye P, Schmitt C, Simian foamy virus transmission from apes to humans, rural Cameroon. Emerg Infect Dis. 2007;13:1314–20.PubMedGoogle Scholar
- Wolfe ND, Heneine W, Carr JK, Garcia AD, Shanmugam V, Tamoufe U, Emergence of unique primate T-lymphotropic viruses among central African bushmeat hunters. Proc Natl Acad Sci U S A. 2005;102:7994–9. DOIPubMedGoogle Scholar
- Wolfe ND, Switzer WM, Carr JK, Bhullar VB, Shanmugam V, Tamoufe U, Naturally acquired simian retrovirus infections in central African hunters. Lancet. 2004;363:932–7. DOIPubMedGoogle Scholar
- Zheng H, Wolfe ND, Sintasath DM, Tamoufe U, Lebreton M, Djoko CF, Emergence of a novel and highly divergent HTLV-3 in a primate hunter in Cameroon. Virology. 2010;401:137–45. DOIPubMedGoogle Scholar
- Van Heuverswyn F, Peeters M. The origins of HIV and implications for the global epidemic. Curr Infect Dis Rep. 2007;9:338–46. DOIPubMedGoogle Scholar
- Aghokeng AF, Liu W, Bibollet-Ruche F, Loul S, Mpoudi-Ngole E, Laurent C, Widely varying SIV prevalence rates in naturally infected primate species from Cameroon. Virology. 2006;345:174–89. DOIPubMedGoogle Scholar
- Aghokeng AF, Ayouba A, Mpoudi-Ngole E, Loul S, Liegeois F, Delaporte E, Extensive survey on the prevalence and genetic diversity of SIVs in primate bushmeat provides insights into risks for potential new cross-species transmissions. Infect Genet Evol. 2010;10:386–96. DOIPubMedGoogle Scholar
- Simon F, Souquière S, Damond F, Kfutwah A, Makuwa M, Leroy E, Synthetic peptide strategy for the detection of and discrimination among highly divergent primate lentiviruses. AIDS Res Hum Retroviruses. 2001;17:937–52. DOIPubMedGoogle Scholar
- Ndongmo CB, Switzer WM, Pau CP, Zeh C, Schaefer A, Pieniazek D, New multiple antigenic peptide-based enzyme immunoassay for detection of simian immunodeficiency virus infection in nonhuman primates and humans. J Clin Microbiol. 2004;42:5161–9. DOIPubMedGoogle Scholar
- Bernhard OK, Mathias RA, Barnes TW, Simpson RJ. A fluorescent microsphere-based method for assay of multiple analytes in plasma. Methods Mol Biol. 2011;728:195–206. DOIPubMedGoogle Scholar
- Crowther JR. The ELISA guidebook. In: Walker JM, editor. Methods in molecular biology. Vol. 149. Totowa (NJ): Humana Press: Totowa, NJ; 2001.
- Monleau M, Montavon C, Laurent C, Segondy M, Montes B, Delaporte E, Evaluation of different RNA extraction methods and storage conditions of dried plasma or blood spots for human immunodeficiency virus type 1 RNA quantification and PCR amplification for drug resistance testing. J Clin Microbiol. 2009;47:1107–18. DOIPubMedGoogle Scholar
- Van Der Kuyl AC, Van Gennep DR, Dekker JT, Goudsmit J. Routine DNA analysis based on 12S rRNA gene sequencing as a tool in the management of captive primates. J Med Primatol. 2000;29:307–15. DOIPubMedGoogle Scholar
- Clewley JP, Lewis JC, Brown DW, Gadsby EL. A novel simian immunodeficiency virus (SIVdrl) pol sequence from the drill monkey, Mandrillus leucophaeus. J Virol. 1998;72:10305–9.PubMedGoogle Scholar
- Aghokeng AF, Bailes E, Loul S, Courgnaud V, Mpoudi-Ngole E, Sharp PM, Full-length sequence analysis of SIVmus in wild populations of mustached monkeys (Cercopithecus cephus) from Cameroon provides evidence for two co-circulating SIVmus lineages. Virology. 2007;360:407–18. DOIPubMedGoogle Scholar
- Liégeois F, Lafay B, Formenty P, Locatelli S, Courgnaud V, Delaporte E, Full-length genome characterization of a novel simian immunodeficiency virus lineage (SIVolc) from olive Colobus (Procolobus verus) and new SIVwrcPbb strains from Western Red Colobus (Piliocolobus badius badius) from the Tai Forest in Ivory Coast. J Virol. 2009;83:428–39. DOIPubMedGoogle Scholar
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. DOIPubMedGoogle Scholar
- Milne I, Lindner D, Bayer M, Husmeier D, McGuire G, Marshall DF, TOPALi v2: a rich graphical interface for evolutionary analyses of multiple alignments on HPC clusters and multi-core desktops. Bioinformatics. 2009;25:126–7. DOIPubMedGoogle Scholar
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. DOIPubMedGoogle Scholar
- Groves C. Primate taxonomy. Smithsonian Series in Comparative Evolutionary Biology. Washington: Smithsonian Institution Press; 2001.
- Goldberg TL, Sintasath DM, Chapman CA, Cameron KM, Karesh WB, Tang S, Coinfection of Ugandan red colobus (Procolobus [Piliocolobus] rufomitratus tephrosceles) with novel, divergent delta-, lenti-, and spumaretroviruses. J Virol. 2009;83:11318–29. DOIPubMedGoogle Scholar
- Verschoor EJ, Fagrouch Z, Bontjer I, Niphuis H, Heeney JL. A novel simian immunodeficiency virus isolated from a Schmidt’s guenon (Cercopithecus ascanius schmidti). J Gen Virol. 2004;85:21–4. DOIPubMedGoogle Scholar
- Takemura T, Ekwalanga M, Bikandou B, Ido E, Yamaguchi-Kabata Y, Ohkura S, A novel simian immunodeficiency virus from black mangabey (Lophocebus aterrimus) in the Democratic Republic of Congo. J Gen Virol. 2005;86:1967–71. DOIPubMedGoogle Scholar
- Aghokeng AF, Ayouba A, Ahuka S, Liegois F, Mbala P, Muyembe JJ, Genetic diversity of simian lentivirus in wild De Brazza’s monkeys (Cercopithecus neglectus) in equatorial Africa. J Gen Virol. 2010;91:1810–6. DOIPubMedGoogle Scholar
- Parrish CR, Holmes EC, Morens DM, Park EC, Burke DS, Calisher CH, Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev. 2008;72:457–70. DOIPubMedGoogle Scholar
- Santiago ML, Range F, Keele BF, Li Y, Bailes E, Bibollet-Ruche F, Simian immunodeficiency virus infection in free-ranging sooty mangabeys (Cercocebus atys atys) from the Tai Forest, Côte d'Ivoire: implications for the origin of epidemic human immunodeficiency virus type 2. J Virol. 2005;79:12515–27. DOIPubMedGoogle Scholar
- Switzer W, Ahuka-Mundeke S, Tang S, Shankar A, Wolfe N, Heneine W, SFV infection from Colobus monkeys in women from Democratic Republic of Congo. 18th Conference on Retroviruses and Opportunistic Infections, Boston, Feb 27−March 2, 2011. Paper no. 454.
- US Agency for International Development HIV/AIDS. Democratic Republic of Congo [cited 2011 Sep 28].) http://www.usaid.gov/our_work/global_health/aids/Countries/africa/congo.html
- Vidal N, Peeters M, Mulanga-Kabeya C, Nzilambi N, Robertson D, Ilunga W, Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J Virol. 2000;74:10498–507. DOIPubMedGoogle Scholar
- Braeckman C. Les nouveaux predateurs. Politique des puissances en Afrique centrale. Paris: Fayard; 2003.
- Laurent C, Bourgeois A, Mpoudi M, Butel C, Peeters M, Mpoudi-Ngolé E, Commercial logging and HIV epidemic, rural Equatorial Africa. Emerg Infect Dis. 2004;10:1953–6.PubMedGoogle Scholar
- Mulanga C, Bazepeo SE, Mwamba JK, Butel C, Tshimpaka JW, Kashi M, Political and socioeconomic instability: how does it affect HIV? A case study in the Democratic Republic of Congo. AIDS. 2004;18:832–4. DOIPubMedGoogle Scholar
Page created: November 30, 2011
Page updated: November 30, 2011
Page reviewed: November 30, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.