Volume 18, Number 3—March 2012
Dispatch
Clinical Significance of Escherichia albertii
Abstract
Discriminating Escherichia albertii from other Enterobacteriaceae is difficult. Systematic analyses showed that E. albertii represents a substantial portion of strains currently identified as eae-positive Escherichia coli and includes Shiga toxin 2f–producing strains. Because E. albertii possesses the eae gene, many strains might have been misidentified as enterohemorrhagic or enteropathogenic E. coli.
Attaching and effacing pathogens possess a locus of enterocyte effacement (LEE)–encoded type III secretion system. They form attaching and effacing lesions on intestinal epithelial cell surfaces by the combined actions of intimin, an eae gene–encoded outer membrane protein, and type III secretion system effectors. Attaching and effacing pathogens include enterohemorrhagic and enteropathogenic Escherichia coli (EHEC and EPEC, respectively) and Citrobacter rodentium (1,2). Escherichia albertii have recently been added to this group (3–5). However, the clinical significance of E. albertii has yet to be fully elucidated, partly because it is difficult to discriminate E. albertii from other Enterobacteriaceae spp. by using routine bacterial identification systems based on biochemical properties (6–9). A large number of E. albertii strains might have been misidentified as EPEC or EHEC because they possess the eae gene.
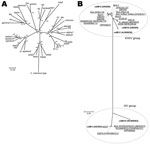
We collected 278 eae-positive strains that were originally identified by routine diagnostic protocols as EPEC or EHEC. They were isolated from humans, animals, and the environment in Japan, Belgium, Brazil, and Germany during 1993–2009 (Table 1; Technical Appendix). To characterize the strains, we first determined their intimin subtypes by sequencing the eae gene as described (Technical Appendix). Of the 275 strains examined, 267 possessed 1 of the 26 known intimin subtypes (4 subtypes—η, ν, τ, and a subtype unique to C. rodentium—were not found). In the remaining 8 strains, we identified 5 new subtypes; each showed <95% nt sequence identity to any known subtype, and they were tentatively named subtypes N1–N5. For subtype N1, 3 variants were identified (N1.1, N1.2, and N1.3, with >95% sequence identity among the 3 variants) (Figure 1, panel A).
To determine the phylogenetic relationships of the strains, we performed multilocus sequencing analysis of 179 strains that were selected from our collection on the basis of intimin subtype and serotype (see Technical Appendix for selection criteria and analysis protocol). Among the 179 strains, 26 belonged to the E. albertii lineage (Figure 2). The 26 E. albertii strains were from 14 humans (13 from symptomatic patients), 11 birds, and 1 cat. All of the 5 new intimin subtypes were found in the E. albertii strains. Intimin subtypes found in other E. albertii strains were also rare subtypes found in E. coli (10). This finding suggests that more previously unknown intimin subtypes may exist in the E. albertii population.
We next analyzed the pheV, selC, and pheU loci of the 26 E. albertii strains for the presence of LEE elements as described (Technical Appendix). These 3 genomic loci are the known LEE integration sites in E. coli. By this analysis, all E. albertii strains except 1 (EC05–44) contained the LEE in the pheU locus (the integration site in EC05–44 was not identified). This finding indicates that despite the remarkable diversity of intimin subtypes, the LEE elements are preferentially integrated into the pheU tRNA gene in E. albertii.
Because all E. albertii strains isolated so far contained the cdtB gene encoding the cytolethal distending toxin B subunit (8,9), we examined the presence and subtype of the cdtB gene as described (Technical Appendix). This analysis revealed that all E. albertii strains except 1 (CB10113) possessed the cdtB gene belonging to the II/III/V subtype group (Figure 1, panel B); this finding is consistent with published findings (9). In addition, 2 strains (E2675 and HIPH08472) each of which was subtype I , possessed a second cdtB gene, (Figure 1, panel B).
We used PCR to further investigate the presence of Shiga toxin genes (stx) and their variants (Technical Appendix) and found that 2 E. albertii strains possessed the stx2f gene (Figure 2, panel B). Stx2 production by these strains was confirmed by using a reverse-passive latex agglutination kit (Technical Appendix). The 2 stx2f-positive strains were those containing the subtype I cdtB gene in addition to the II/III/V subtype group gene: 1 (HIPH08472) was isolated from a patient with diarrhea and the other (E2675) was from a healthy Corvus sp. bird (Figure 2).
Last, we examined the phenotypic and biochemical properties of the 26 E. albertii strains and compared the results with those obtained in a previous study (9) and with those of E. albertii type strain LMG20976 (Table 2). To identify features that could discriminate E. albertii from E. coli, the results were further compared with those of E. coli (11). Consistent with findings in previous reports (5–7,9), the lack of motility and the inability to ferment xylose and lactose and to produce β-D-glucuronidase were common biochemical properties of E. albertii that could be used to discriminate E. albertii from E. coli, although 1 E. albertii strain was positive for lactose fermentation. The inability of E. albertii to ferment sucrose has been described as a common feature (9); however, a positive reaction to this test was found for 5 (19.2%) E. albertii strains. Moreover, approximately half of E. coli strains are positive for sucrose fermentation. Thus, the inability to ferment sucrose is not informative. Rather, the inability to ferment dulcitol (all E. albertii strains were negative, 60% of E. coli strains are positive) may be a useful biochemical property for differentiation.
In the current clinical laboratory setting, a substantial number of E. albertii strains are misidentified as EPEC or EHEC. Because 13 of the isolates were from patients with signs and symptoms of gastrointestinal infection, E. albertii is probably a major enteric human pathogen. In addition, E. albertii should be regarded as a potential Stx2f-producing bacterial species, although the clinical significance of Stx2f-producing strains is unknown.
Notable genetic, phenotypic, and biochemical properties of E. albertii, which were identified by analyzing the confirmed E. albertii strains, are 1) possession of intimin subtypes rarely or previously undescribed in E. coli, 2) possession of the II/III/V subtype group cdtB gene, 3) LEE integration into the pheU tRNA gene, 4) nonmotility, and 5) inability to ferment xylose, lactose, and dulcitol (but not sucrose) and to produce β-D-glucuronidase. These properties could be useful for facilitating identification of E. albertii strains in clinical laboratories, which would in turn improve understanding of the clinical significance and the natural host and niche of this newly recognized pathogen. In this regard, however, current knowledge of the genetic and biological properties of E. albertii might be biased toward a certain group of E. albertii strains because, even with this study, only a limited number of strains have been analyzed. To more precisely understand the properties of E. albertii as a species, further analysis of more strains from various sources is necessary.
Dr Ooka is an assistant professor at the Department of Infectious Diseases, Faculty of Medicine, University of Miyazaki. His research interests include the genomics and pathogenicity of pathogenic bacteria, especially attaching and effacing pathogens.
Acknowledgment
This work was supported by the following KAKENHI (Grants-in-Aid for Scientific Research) grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan: Applied Genomics, 17019058, to T.H.; Kiban-B, 20310116, to T.H.; and Wakate-B, 23790480, to T.O.
References
- Croxen MA, Finlay BB. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol. 2010;8:26–38.PubMedGoogle Scholar
- Schmidt MA. LEEways: tales of EPEC, ATEC and EHEC. Cell Microbiol. 2010;12:1544–52. DOIPubMedGoogle Scholar
- Albert MJ, Alam K, Islam M, Montanaro J, Rahman AS, Haider K, Hafnia alvei, a probable cause of diarrhea in humans. Infect Immun. 1991;59:1507–13.PubMedGoogle Scholar
- Albert MJ, Faruque SM, Ansaruzzaman M, Islam MM, Haider K, Alam K, Sharing of virulence-associated properties at the phenotypic and genetic levels between enteropathogenic Escherichia coli and Hafnia alvei. J Med Microbiol. 1992;37:310–4. DOIPubMedGoogle Scholar
- Huys G, Cnockaert M, Janda JM, Swings J. Escherichia albertii sp. nov., a diarrhoeagenic species isolated from stool specimens of Bangladeshi children. Int J Syst Evol Microbiol. 2003;53:807–10. DOIPubMedGoogle Scholar
- Janda JM, Abbott SL, Albert MJ. Prototypal diarrheagenic strains of Hafnia alvei are actually members of the genus Escherichia. J Clin Microbiol. 1999;37:2399–401.PubMedGoogle Scholar
- Abbott SL, O'Connor J, Robin T, Zimmer BL, Janda JM. Biochemical properties of a newly described Escherichia species, Escherichia albertii. J Clin Microbiol. 2003;41:4852–4. DOIPubMedGoogle Scholar
- Hyma KE, Lacher DW, Nelson AM, Bumbaugh AC, Janda JM, Strockbine NA, Evolutionary genetics of a new pathogenic Escherichia species: Escherichia albertii and related Shigella boydii strains. J Bacteriol. 2005;187:619–28. DOIPubMedGoogle Scholar
- Oaks JL, Besser TE, Walk ST, Gordon DM, Beckmen KB, Burek KA, Escherichia albertii in wild and domestic birds. Emerg Infect Dis. 2010;16:638–46 .DOIPubMedGoogle Scholar
- Blanco M, Schumacher S, Tasara T, Zweifel C, Blanco JE, Dahbi G, Serotypes, intimin variants and other virulence factors of eae positive Escherichia coli strains isolated from healthy cattle in Switzerland. Identification of a new intimin variant gene (eae-eta2). BMC Microbiol. 2005;5:23. DOIPubMedGoogle Scholar
- Nataro JP, Bopp CA, Fields PI, Kaper JB, Strockbine NA. Escherichia, Shigella, and Salmonella. In: Murray PR, Baron EJ, Jorgensen JH, Landry ML, Pfaller MA, editors. Manual of clinical microbiology, 9th ed. Washington: ASM Press; 2007. p. 670–87.
Figures
Tables
Cite This ArticleTable of Contents – Volume 18, Number 3—March 2012
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Tetsuya Hayashi, Division of Bioenvironmental Science, Frontier Science Research Center, University of Miyazaki, Kihara 5200, Kiyotake, Miyazaki 889-1692, Japan
Top