Volume 18, Number 3—March 2012
Research
Causes of Pneumonia Epizootics among Bighorn Sheep, Western United States, 2008–2010
Abstract
Epizootic pneumonia of bighorn sheep is a devastating disease of uncertain etiology. To help clarify the etiology, we used culture and culture-independent methods to compare the prevalence of the bacterial respiratory pathogens Mannheimia haemolytica, Bibersteinia trehalosi, Pasteurella multocida, and Mycoplasma ovipneumoniae in lung tissue from 44 bighorn sheep from herds affected by 8 outbreaks in the western United States. M. ovipneumoniae, the only agent detected at significantly higher prevalence in animals from outbreaks (95%) than in animals from unaffected healthy populations (0%), was the most consistently detected agent and the only agent that exhibited single strain types within each outbreak. The other respiratory pathogens were frequently but inconsistently detected, as were several obligate anaerobic bacterial species, all of which might represent secondary or opportunistic infections that could contribute to disease severity. These data provide evidence that M. ovipneumoniae plays a primary role in the etiology of epizootic pneumonia of bighorn sheep.
In North America, epizootic pneumonia is a devastating, population-limiting disease of bighorn sheep (Ovis canadensis) (1–5). Anecdotal and experimental evidence suggests that in at least some instances, this disease may be introduced into bighorn sheep populations by contact with domestic sheep or goats (5,6). When the disease is first introduced, outbreaks affect animals of all ages (1–3). During subsequent years or decades, sporadic cases of pneumonia in adult sheep and annual epizootics of pneumonia in lambs may continue (7–10).
Considering the dramatic and severe character of epizootic bighorn sheep pneumonia, the etiology is surprisingly unclear. Findings of gross and histopathologic examinations of lung tissue strongly suggest bacterial etiology: anterior–ventral distribution, suppurative inflammation, and abundant bacterial colonies. In domestic ruminants, bacterial pneumonia frequently occurs secondary to viral infections or other pulmonary insults, but extensive efforts to detect such underlying factors for bighorn sheep pneumonia have generally been nonproductive. For example, although evidence of infection or exposure to respiratory viruses, especially respiratory syncytial virus and parainfluenza virus, is frequently found in healthy and pneumonia-affected populations, no consistent association between the disease and any virus has been found (11–13). As a result, most research attention has been directed toward bacterial respiratory pathogens that may act as primary infectious agents, particularly leukotoxin-expressing Mannheimia haemolytica, which is highly lethal to bighorn sheep after experimental challenge (5,14). Other Pasteurellaceae, particularly Bibersteinia trehalosi and Pasteurella multocida, have been more frequently isolated from pneumonia-affected animals during natural outbreaks than has M. haemolytica (11,12,15). Another candidate pathogen, Mycoplasma ovipneumoniae, has recently been isolated from pneumonia-affected bighorn sheep during 2 epizootics (11,16,17); antibodies against this agent were detected in bighorn sheep from 9 populations undergoing pneumonia epizootics but were absent in 9 nonaffected populations (17). In experiments, M. ovipneumoniae has been shown to predispose bighorn sheep to M. haemolytica pneumonia (18). When M. ovipneumoniae–free domestic sheep were commingled with bighorn sheep, the bighorn sheep survived at unprecedented rates (19).
Development of effective methods for managing, preventing, or treating an infectious disease requires a clear understanding of its underlying etiology. However, clarifying the etiology can be difficult, particularly for primary infections (e.g., HIV) that are characteristically associated with multiple opportunistic infections that may be more lethal than the epidemic agent itself. During 2008–2010, epizootic pneumonia of bighorn sheep was detected in at least 5 western US states. These epizootics provided an opportunity to conduct a comparative study of the etiology of this disease (Table 1).
Conventional microbiological methods can fail to isolate agents because of their fastidious in vitro growth requirements or intermicrobial interactions; thus, for agent isolation, we used 2 culture-independent methods (agent-specific PCRs and 16S clone libraries) in addition to conventional bacterial cultures (17,20–22). We expected that primary etiologic agents could be differentiated from opportunistic agents by 1) their detection at high prevalence in affected animals, 2) the presence of single (clonal) strain types within each outbreak, and 3) their uncommon or lack of detection in animals from healthy populations (11,22–24). Therefore, to clarify the etiology of epizootic pneumonia, we applied these criteria to the bacterial respiratory pathogens detected in multiple bighorn sheep epizootics.
Bighorn Sheep Populations
The study sample consisted of 8 demographically independent bighorn sheep populations in 5 states (Montana, Nevada, Washington, Oregon, and South Dakota) that had been affected by epizootic pneumonia during 2008–2010 and for which lung tissue specimens from >4 affected animals were available (Table 1). In 6 of these populations, the disease affected bighorn sheep of all ages; in the other 2 populations, in which the disease had previously affected sheep of all ages, the disease was restricted to lambs. Convenience samples were selected among those available from each epizootic: the sample of pneumonia-affected animals consisted of the first 4–6 sheep for which pneumonia had been confirmed by gross or microscopic lesions. Sheep initially selected for analysis but later determined to have lacked gross or microscopic lesions characteristic of pneumonia were retained in the study but analyzed separately. Negative controls consisted of animals with no gross or histopathologic evidence of pneumonia that died or were culled from 2 closely observed healthy populations.
Bacteriologic Cultures
Surfaces of affected lung tissue specimens were seared, and swab samples of deeper tissues were obtained and streaked onto Columbia blood agar plates (Hardy Diagnostics, Santa Maria, CA, USA). Pasteurellaceae were isolated and identified by using routine methods (25) and then stored at −80°C in 30% buffered glycerol in brain–heart infusion agar (Hardy Diagnostics).
DNA Template Preparation
DNA was extracted from 1.0–1.5 mL of fluid collected from 1–2 g of fresh-frozen lung tissue macerated in 1 mL of phosphate buffered saline for 5 min by using a stomacher (Seward Stomacher 80 Laboratory Blender, Bohemia, NY, USA). DNA was extracted by using a QIAamp mini kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s protocol.
PCR Detection of Respiratory Pathogens and lktA
To detect M. haemolytica, P. multocida, B. trehalosi, lktA, and M. ovipneumoniae, we used previously published PCR protocols with minor modifications (Table 2). All reactions were conducted individually in 20-μL volumes containing 2 μL of DNA template (5–1,000 ng/μL), 10 μL of master mix (QIAGEN Hotstar mix for P. multocida, M. ovipneumoniae, and lktA and QIAGEN Multiplex PCR mix for B. trehalosi and M. haemolytica), and primers at 0.2 μmol (P. multocida, M. haemolytica, and B. trehalosi), 2 μmol (M. ovipneumoniae), or 0.5 μmol (lktA). Thermocycler conditions included an initial denaturation step at 95°C (15 min) for all agents and a final extension step at 72°C (5 min, except final extensions for P. multocida and lktA were 9 and 10 min, respectively). Cycling conditions used were as follows: for M. ovipneumoniae, 30 cycles at 95°C for 30 s, at 58°C for 30 s, and at 72°C for 30 s; for B. trehalosi and M. haemolytica, 35 cycles at 95°C for 30 s, at 55°C for 30 s, and at 72°C for 40 s; and for P. multocida and lktA, 30 cycles at 95°C for 60 s, at 55°C for 60 s, and at 72°C for 60 s. Amplicons were examined in UV light after electrophoresis in 1.2% agarose gel containing 0.005% ethidium bromide in 0.5× Tris/borate/EDTA buffer at 7 V/cm.
16S Analyses
To detect predominant microbial populations in the pneumonic lung tissues, we used a culture-independent method (17). In brief, we aseptically collected two 1-g samples of lung tissue from sites at least 10 cm apart in grossly abnormal (consolidated) tissue from 16 pneumonia-affected animals, including 2 from each outbreak. Tissues were stomached and DNA was extracted (DNeasy Blood and Tissue Kit; QIAGEN) from 100-μg aliquots of each homogenate. Segments of 16S rDNA were PCR amplified and cloned. Insert DNA was sequenced (vector primers T3 and M13, BigDye version 3.1, ABI PRISM Genetic Analyzer; Applied Biosystems, Foster City, CA, USA) from 16 clones derived from each homogenate, resulting in 32 sequences from each animal. DNA sequences were assigned to species (>99% identity) or genus (>97% identity) according to BLASTN GenBank search results (29). Clone sequences may be accessed in GenBank under accession nos. JN857366–857894.
Pulsed-Field Gel Electrophoresis for Pasteurellaceae
When available, Pasteurellaceae isolated from the study animals were obtained from the Washington Animal Disease Diagnostic Laboratory (Pullman, WA, USA). If such isolates were unavailable, we substituted banked isolates from other bighorn sheep involved in the same outbreaks. Isolates were subjected to pulsed-field gel electrophoresis (PFGE) performed on a CHEF-DRII PFGE apparatus (Bio-Rad, Hercules, CA, USA) in 1% agarose gel (Seakem Gold Agarose; FMC Bio Products, Rockland, MD, USA) in 0.5× Tris borate EDTA buffer at 14°C for 20 h at 6 V/cm and a linear ramp of 1.0–30.0 s for ApaI or 0.5–40.0 s for SmaI. Salmonella serovar Braenderup H9812, digested with XbaI for 3 h at 37°C, was used as a size standard on each gel. Gels were stained with ethidium bromide and photographed under UV transillumination. PFGE data were analyzed by using BioNumerics version 4.6 (www.applied-maths.com/bionumerics/bionumerics.htm) with Dice coefficients and the unpaired pair group method with arithmetic means for clustering; tolerance and optimization parameters were set at 1%.
Intergenic Spacer Region Sequence typing for M. ovipneumoniae
In a preliminary study performed in our laboratory, ribosomal operon intergenic spacer (IGS) regions of M. ovipneumoniae from isolates from 6 bighorn sheep populations were amplified by using the method described by Kong et al. (30) and sequenced (Amplicon Express, Pullman, WA, USA). Sequences were aligned and clustered by using Lasergene software (DNASTAR, Inc., Madison WI, USA). Each isolate exhibited a different IGS sequence, demonstrating the utility of IGS sequences for identifying strain diversity (data not shown). We used Primer3 software (http://frodo.wi.mit.edu/primer3/) to develop primers flanking the variable IGS region, conserved among M. ovipneumoniae isolates, and divergent from IGS regions of the second most common sheep upper respiratory mycoplasma, M. arginini (Table 2). IGS PCR products were sequenced, and sequences were aligned and clustered by using Lasergene software. IGS sequences can be accessed in GenBank under accession nos. JN857895–857934.
Statistical Analyses
To evaluate the agreement between results of bacteriologic cultures and PCR tests for detection of P. multocida, M. haemolytica, and B. trehalosi, we used Cohen κ coefficients (31). To assess overall differences in prevalence of specific bacterial respiratory pathogens, we used χ2 tests; for pairwise comparisons, we used the Marascuilo procedure (32) to control for multiple comparison problems, which might affect error rates. To assess associations between prevalence of different respiratory bacteria and mortality rates among different bighorn sheep populations, we used the Pearson correlation coefficient.
We detected 4 previously reported bacterial respiratory pathogens of bighorn sheep. We detected M. haemolytica, B. trehalosi, and P. multocida by using aerobic culture and species-specific PCR and M. ovipneumoniae by using PCR alone (20) (Table 3; Table A1). Agreement between detection by culture and PCR varied by agent, ranging from no agreement (M. haemolytica, κ −0.02), to fair agreement (B. trehalosi, κ 0.22), to good agreement (P. multocida, κ 0.76). For the purposes of the following analyses, animals for which any agent was detected by either method were considered positive for that agent. Among the targeted agents, 3 (B. trehalosi, M. haemolytica, and M. ovipneumoniae) were detected in >1 animals from all 8 outbreak-affected populations and 1 (P. multocida) was detected in animals from 5 outbreak-affected populations (Table 3).
Frequency of detecting M. haemolytica, B. trehalosi, P. multocida, and M. ovipneumoniae from pneumonia-affected animals differed significantly (χ2 26.2, 3 df, p<0.0001). M. ovipneumoniae (n = 42, 95%) was detected significantly more often than any other agent except B. trehalosi (n = 35, 73%; Marascuilo procedure, p<0.05). Detection of lktA, a gene encoding the leukotoxin expressed by B. trehalosi and M. haemolytica, was then analyzed as a surrogate for virulent M. haemolytica and/or B. trehalos because these species, if lacking lktA, would be considered to have greatly reduced or no virulence (33). Prevalence of lktA (n = 10, 22.7%) was significantly lower than that of P. multocida (n = 21, 47.7%, Marascuilo procedure, p<0.05).
Frequency of detecting B. trehalosi and P. multocida differed significantly among outbreaks (p = 0.002 and 0.001, respectively). Similarly, PCR-based detection of lktA differed among outbreaks (p = 0.003). Although such differences could potentially contribute to the significant differences in disease severity and mortality rates among the epizootics in this study (χ2184.7, 7 df, p<0.0001), the prevalence of B. trehalosi, P. multocida, or lktA did not correlate with estimated mortality rates in the 8 outbreaks included in this study (Tables 1, 3).
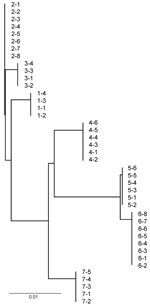
Strain typing to evaluate the genetic similarity of bacterial pathogens within and among outbreaks (23) detected only single IGS types of M. ovipneumoniae within each outbreak, whereas distinctly different IGS types were found for each epizootic with the exception of 2 populations in Montana (Figure). In contrast, the PFGE strain types of Pasteurellaceae isolated from within single outbreaks ranged from 0 to 7, including 0–7 B. trehalosi strains and 0–2 P. multocida strains (Table 4; Table A1). Assessment of strain type diversity of M. haemolytica within outbreaks was not possible because this species was isolated only 1 time.
Among the agents and genes tested, M. ovipneumoniae was the only agent or gene that was detected at different frequencies for animals from epizootic-affected compared with non–epizootic-affected populations (Table 3; p<0.001). The frequency of M. ovipneumoniae and P. multocida detection in non–pneumonia-affected animals culled from epizootic populations was intermediate, significantly lower than that in pneumonia-affected animals (p<0.01).
Partial 16S ribosomal DNA sequences of 527 clones, including ≈30 clones from each of 2 animals from each epizootic, detected the targeted respiratory pathogens P. multocida, M. ovipneumoniae, B. trehalosi, and M. haemolytica, although the latter accounted for <1% of the identifications. Also predominant were several obligate anaerobic bacteria (Fusobacterium necrophorum, Prevotella spp., Clostridium spp., and Bacteroides spp.) (Table 5).
The results of this study support the hypothesis that M. ovipneumoniae is a primary agent in the etiology of epizootic bighorn sheep pneumonia in populations across the western United States and that it acts to induce secondary infection with opportunistic pathogens. M. ovipneumoniae was detected in the pneumonic lungs of >95% of study animals involved in the 8 discrete pneumonia epizootics, significantly more frequently than any of the other respiratory agents sought except the bighorn sheep commensal bacterium B. trehalosi (34,35). We identified identical ribosomal IGS strains of M. ovipneumoniae within the affected animals in each outbreak, consistent with epizootic spread (24); M. ovipneumoniae was not detected in the healthy populations sampled. Of note, the 2 populations in which identical IGS strains of M. ovipneumoniae were detected were separated by only ≈20 miles, suggesting the possibility that this strain was transmitted among these populations by movement of >1 M. ovipneumoniae–infected bighorn sheep.
The normal host range of M. ovipneumoniae (members of Old World Caprinae, including domestic sheep and mouflon, a closely related Eurasian sheep species) is consistent with many observations that epizootic bighorn sheep pneumonia frequently follows contact with these hosts (5,19). Previous experiments in which bighorn sheep were commingled with domestic sheep or mouflon each resulted in epizootic bighorn sheep pneumonia and, cumulatively, the death of 88 (98%) of 90 bighorn sheep; similar commingling experiments with other ungulates (deer, elk, llamas, horses, cattle, goats, mountain goats) resulted at most in sporadic deaths from bighorn sheep pneumonia (4 [7%] of 56) (19). In a recent study in which bighorn sheep were commingled with M. ovipneumoniae–free domestic sheep, the lack of epizootic bighorn sheep pneumonia was unprecedented (19). Together, these data support the hypothesis that bighorn sheep epizootic pneumonia results from cross-species transmission of M. ovipneumoniae from its normal host(s) to a naive, highly susceptible host: bighorn sheep.
Each of the other specific potential respiratory pathogens targeted failed to fulfill >1 expectations for a primary etiologic agent. B. trehalosi was detected in most animals regardless of their health status; exhibited diverse strain types within epizootics; and in most instances was detected in the absence of lktA, consistent with the nontoxigenic strains widely distributed in healthy and pneumonic bighorn sheep (36). M. haemolytica was similarly detected in about half of the animals regardless of their health status and in the absence of lktA. P. multocida was not detected at all in animals involved in 3 of the epizootics, but in those outbreaks in which it was present, it was detected at high prevalence and somewhat more frequently in pneumonia-affected than in healthy bighorn sheep. Furthermore, multiple isolates from those epizootics in which it was detected shared a high degree of genetic similarity, consistent with epizootic transmission (24).
The frequencies with which B. trehalosi, P. multocida, and lktA were detected from animals in the different epizootics differed significantly, although this finding did not correlate with mortality rates (Table 1). This conclusion is limited, however, by the possible confounding effect of the extensive culling conducted in several areas of the epizootics examined in this study. More research into factors that affect the severity of bighorn sheep pneumonia epizootics is clearly needed.
The analysis of prevalence of bacterial respiratory pathogens in the lung tissues of healthy animals from unaffected populations was comparatively limited by the small number of control specimens available. To more clearly define the prevalence, infectivity, and virulence of M. ovipneumoniae, sampling of additional healthy bighorn sheep populations is needed. Although M. ovipneumoniae was not detected in the negative control animals examined in this study and although serologic evidence of exposure to M. ovipneumoniae is uncommon or rare in healthy bighorn sheep populations (17), several apparently healthy bighorn sheep populations with serologic and/or PCR evidence of exposure to M. ovipneumoniae have been identified (data not shown). This finding demonstrates that not all exposures to this agent result in epizootic bronchopneumonia or, perhaps, that unrecognized previous epizootics had occurred. To clarify these findings, more research, specifically including longitudinal observational studies and investigation of strain differences in virulence of M. ovipneumoniae (37,38), is needed.
Our universal eubacterial 16S rDNA approach used analysis of small clone libraries from each animal to detect those agents representing >10% of the 16S operons in lung tissue with high (>95%) confidence. The 3 most frequently detected aerobic bacterial agents detected by using this method were P. multocida, M. ovipneumoniae, and B. trehalosi; detection of M. haemolytica was rare (0.19%) despite the high frequency of its detection by the more sensitive PCR. This finding is surprising because M. haemolytica has been regarded as the principal pneumonia pathogen of bighorn sheep (5). It has been argued that because B. trehalosi inhibits or kills M. haemolytica in coculture, the same effect in vivo may block detection of M. haemolytica in bighorn sheep lungs (21), but this argument cannot explain the dearth of M. haemolytica detected in this study because the proportion of lungs that were positive for M. haemolytica by PCR was actually lower in the absence of B. trehalosi (Table A1). Furthermore, the lung tissues from animals affected by the 5 epizootics in Washington or Montana were obtained from bighorn sheep that were coughing and culled in an attempt to prevent further transmission of the disease; therefore, these specimens could represent animals at earlier stages of the disease when more consistent presence of causal agents would be expected.
Consistent with previous reports of bighorn sheep in Hells Canyon (17), the predominance of obligate anaerobes (Fusobacterium, Prevotella, Clostridium, and Bacteroides spp.) among the lung flora was consistent with decreased clearance of inhaled oral flora from the lower respiratory tract. Impaired clearance of inhaled flora is expected subsequent to infection by virulent M. ovipneumoniae (38) or by leukotoxin-expressing Pasteurellaceae (39), albeit by different mechanisms.
To our knowledge, only 1 other study of epizootic bighorn sheep pneumonia has reported comparative microbiological findings from pneumonia-affected animals involved in multiple discrete epizootics. Aune et al. (12) reported that Pasteurellaceae cultured from pneumonia-affected animals differed somewhat among 4 bighorn sheep pneumonia epizootics in Montana during 1991–1996. P. multocida was isolated from pneumonic lung tissues of >1 animals during all 4 epizootics, although prevalence exceeded 50% during only 1 epizootic. Pasteurellaceae biotypes corresponding to B. trehalosi were isolated from animals involved in 3 of the 4 outbreaks, and Pasteurellaceae biotypes corresponding to M. haemolytica were isolated from animals in only 1 outbreak. The microbiology of epizootic pneumonia in Hells Canyon also has been described (11,15,40); results were broadly comparable to the conventional microbiology results reported here for Pasteurellaceae. All these studies differed from the study reported here in that the conventional microbiological methods used failed to recognize M. ovipneumoniae in affected lung tissues.
In summary, of the bacterial respiratory pathogens evaluated, M. ovipneumoniae was the only agent for which the data consistently met the criteria for a primary etiologic agent across all outbreaks. In contrast, the data were inconsistent with regard to a primary etiologic role for any Pasteurellaceae species. The likelihood of M. ovipneumoniae having a primary role in bighorn sheep pneumonia is consistent with the association between some epizootics of this disease and contact with domestic sheep because the latter carry this agent at high prevalence. Identification of M. ovipneumoniae as the epizootic agent of bighorn sheep pneumonia may provide a useful focus for the development of specific preventative or therapeutic interventions.
Dr Besser is a professor of veterinary microbiology at Washington State University and the Washington Animal Disease Diagnostic Laboratory. His research interests include preharvest food safety microbiology and infectious diseases of animals.
Acknowledgments
We thank those persons whose work made this project possible, including Victoria L. Edwards, Craig Jourdonnais, Ray Vinkey, Michael Thompson, Keri Carson, Jeff Bernatowicz, Mark Vekasy, Roblyn Brown, Caleb McAdoo, Kari Huebner, Jeremy Lutz, Scott Roberts, Ken Gray, Chris Morris, Daniel Crowell, Steve Griffin, John Broecher, and numerous biologists from the US Department of Agriculture, Animal Plant Health Inspection Service, Wildlife Services National Wildlife Disease Program.
Funding for this project was provided by the Washington Department of Fish and Wildlife; the Nevada Department of Wildlife; Nevada Bighorns Unlimited; the Oregon Department of Fish and Wildlife; the Idaho Department of Fish and Game; the Idaho Wildlife Disease Research Oversight Committee; Federal Aid in Wildlife Restoration (study No. 7537), administered through the South Dakota Department of Game, Fish and Parks; and Project W-160-R, administered through the Idaho Department of Fish and Game.
References
- Cassirer EF, Sinclair ARE. Dynamics of pneumonia in a bighorn sheep metapopulation. J Wildl Manage. 2007;71:1080–8. DOIGoogle Scholar
- Hobbs NT, Miller MW. Interactions between pathogens and hosts: simulation of pasteurellosis epidemics in bighorn sheep populations. In: McCullough DR, Barrett RH, editors. Wildlife 2001: populations. New York: Springer Publishing Company; 1992. p. 997–1007.
- McCarty CW, Miller MW. Modeling the population dynamics of bighorn sheep: a synthesis of literature. Colorado Division of Wildlife special report 73. Denver: Colorado Division of Wildlife; 1998.
- Monello RJ, Murray DL, Cassirer EF. Ecological correlates of pneumonia epizootics in bighorn sheep herds. Can J Zool. 2001;79:1423–32. DOIGoogle Scholar
- Miller MW. Pasteurellosis. In: Williams ES, Barker IK, editors. Infectious diseases of wild mammals. Ames (IA): Iowa State University Press; 2001. p. 558.
- George JL, Martin DJ, Lukacs PM, Miller MW. Epidemic pasteurellosis in a bighorn sheep population coinciding with the appearance of a domestic sheep. J Wildl Dis. 2008;44:388–403.PubMedGoogle Scholar
- Festa-Bianchet M. A pneumonia epizootic in bighorn sheep, with comments on preventive management. In: Samuel WM, editor. Proceedings of the Sixth Biennial Symposium of the Northern Wild Sheep and Goat Council. 1988 Apr 11–15; Banff, Alberta, Canada. Cody (WY): The Council; 1988. p. 66–76.
- Spraker TR, Hibler CP, Schoonveld GG, Adney WS. Pathologic changes and microorganisms found in bighorn sheep during a stress-related die-off. J Wildl Dis. 1984;20:319–27.PubMedGoogle Scholar
- Monello RJ, Murray DL, Cassirer EF. Ecological correlates of pneumonia epizootics in bighorn sheep herds. Can J Zool. 2001;79:1423–32. DOIGoogle Scholar
- Ryder TJ, Mills KW, Bowles KH, Thorne ET. Effect of pneumonia on population size and lamb recruitment in Whiskey Mountain bighorn sheep. In: Proceedings of the Eighth Biennial Symposium of the Northern Wild Sheep and Goat Council; 1992 Apr 27–May 1; Cody, Wyoming, USA. Cody (WY): The Council; 1992. p.136–46.
- Rudolph KM, Hunter DL, Rimler RB, Cassirer EF, Foreyt WJ, DeLong WJ, Microorganisms associated with a pneumonic epizootic in Rocky Mountain bighorn sheep (Ovis canadensis canadensis). J Zoo Wildl Med. 2007;38:548–58. DOIPubMedGoogle Scholar
- Aune KA, Anderson N, Worley D, Stackhouse L, Henderson J, Daniel J. A comparison of population and health histories among seven bighorn sheep populations. Proceedings of the Eleventh Biennial Symposium of the Northern Wild Sheep and Goat Council. 1998 Apr 16–20; Whitefish, Montana, USA; 1998. Cody (WY): The Council; 1998; p.:46–69.
- Clark RK, Jessup DA, Kock MD, Weaver RA. Survey of desert bighorn sheep in California for exposure to selected infectious diseases. J Am Vet Med Assoc. 1985;187:1175–9.PubMedGoogle Scholar
- Foreyt WJ. Fatal Pasteurella haemolytica pneumonia in bighorn sheep after direct contact with clinically normal domestic sheep. Am J Vet Res. 1989;50:341–4.PubMedGoogle Scholar
- Weiser GC, DeLong WJ, Paz JL, Shafii B, Price WJ, Ward ACS. Characterization of Pasteurella multocida associated with pneumonia in bighorn sheep. J Wildl Dis. 2003;39:536–44.PubMedGoogle Scholar
- Wolfe LL, Diamond B, Spraker TR, Sirochman MA, Walsh DP, Machin CM, A bighorn sheep die-off in southern Colorado involving a Pasteurellaceae strain that may have originated from syntopic cattle. J Wildl Dis. 2010;46:1262–8.PubMedGoogle Scholar
- Besser TE, Cassirer EF, Potter KA, VanderSchalie J, Fischer A, Knowles DP, Association of Mycoplasma ovipneumoniae infection with population-limiting respiratory disease in free-ranging Rocky Mountain bighorn sheep (Ovis canadensis canadensis). J Clin Microbiol. 2008;46:423–30. DOIPubMedGoogle Scholar
- Dassanayake RP, Shanthalingam S, Herndon CN, Subramaniam R, Lawrence PK, Bavananthasivam J, Mycoplasma ovipneumoniae can predispose bighorn sheep to fatal Mannheimia haemolytica pneumonia. Vet Microbiol. 2010;145:354–9. DOIPubMedGoogle Scholar
- Besser TE, Cassirer EF, Yamada C, Potter KA, Herndon C, Foreyt WJ, Survival of bighorn sheep (Ovis canadensis) commingled with domestic sheep (Ovis aries) in the absence of Mycoplasma ovipneumoniae. J Wildl Dis. 2012;48:168–72.PubMedGoogle Scholar
- Weiser GC, Drew ML, Cassirer EF, Ward AC. Detection of Mycoplasma ovipneumoniae in bighorn sheep using enrichment culture coupled with genus- and species-specific polymerase chain reaction. J Wildl Dis. 2012. In press.
- Dassanayake RP, Call DR, Sawant AA, Casavant NC, Weiser GC, Knowles DP, Bibersteinia trehalosi inhibits the growth of Mannheimia haemolytica by a proximity-dependent mechanism. Appl Environ Microbiol. 2010;76:1008–13. DOIPubMedGoogle Scholar
- Fredericks DN, Relman DA. Sequence-based identification of microbial pathogens: a reconsideration of Koch's postulates. Clin Microbiol Rev. 1996;9:18–33.PubMedGoogle Scholar
- Gilbert GL. Molecular diagnostics in infectious diseases and public health microbiology: cottage industry to postgenomics. Trends Mol Med. 2002;8:280–7. DOIPubMedGoogle Scholar
- Riley LW. Molecular epidemiology of infectious diseases: principles and practices. Washington: ASM Press; 2004.
- Quinn PJ, Markey BK, Leonard FC, FitzPatrick ES, Fanning S, Hartigan PJ. Veterinary microbiology and microbial disease. 2nd ed. Chichester (UK): Wiley-Blackwell; 2011.
- Fisher MA, Weiser GC, Hunter DL, Ward ACS. Use of a polymerase chain reaction method to detect the leukotoxin gene IktA in biogroup and biovariant isolates of Pasteurella haemolytica and P. trehalosi. Am J Vet Res. 1999;60:1402–6.PubMedGoogle Scholar
- Townsend KM, Frost AJ, Lee CW, Papadimitriou JM, Dawkins HJ. Development of PCR assays for species- and type-specific identification of Pasteurella multocida isolates. J Clin Microbiol. 1998;36:1096–100.PubMedGoogle Scholar
- McAuliffe L, Hatchell FM, Ayling RD, King AI, Nicholas RA. Detection of Mycoplasma ovipneumoniae in Pasteurella-vaccinated sheep flocks with respiratory disease in England. Vet Rec. 2003;153:687–8. DOIPubMedGoogle Scholar
- Petti CA. Detection and identification of microorganisms by gene amplification and sequencing. Clin Infect Dis. 2007;44:1108–14. DOIPubMedGoogle Scholar
- Kong F, James G, Gordon S, Zelynski A, Gilbert GL. Species-specific PCR for identification of common contaminant mollicutes in cell culture. Appl Environ Microbiol. 2001;67:3195–200. DOIPubMedGoogle Scholar
- Cohen J. Weighted kappa: nominal scale agreement with provision for scaled disagreement of partial credit. Psychol Bull. 1968;70:213–20. DOIPubMedGoogle Scholar
- Marascuilo LA. Large-sample multiple comparisons. Psychol Bull. 1966;65:280–90. DOIPubMedGoogle Scholar
- Jeyaseelan S, Sreevatsan S, Maheswaran SK. Role of Mannheimia haemolytica leukotoxin in the pathogenesis of bovine pneumonic pasteurellosis. Anim Health Res Rev. 2002;3:69–82. DOIPubMedGoogle Scholar
- Ward ACS, Hunter DL, Jaworski MD, Benolkin PJ, Dobel MP, Jeffress JB, Pasteurella spp. in sympatric bighorn and domestic sheep. J Wildl Dis. 1997;33:544–57.PubMedGoogle Scholar
- Jaworski MD, Hunter DL, Ward ACS. Biovariants of isolates of Pasteurella from domestic and wild ruminants. J Vet Diagn Invest. 1998;10:49–55. DOIPubMedGoogle Scholar
- Sweeney SJ, Silflow RM, Foreyt WJ. Comparative leukotoxicities of Pasteurella haemolytica isolates from domestic sheep and free-ranging bighorn sheep (Ovis canadensis). J Wildl Dis. 1994;30:523–8.PubMedGoogle Scholar
- Parham K, Churchward CP, McAuliffe L, Nicholas RA, Ayling RD. A high level of strain variation within the Mycoplasma ovipneumoniae population of the UK has implications for disease diagnosis and management. Vet Microbiol. 2006;118:83–90. DOIPubMedGoogle Scholar
- Alley MR, Ionas G, Clarke JK. Chronic non-progressive pneumonia of sheep in New Zealand–a review of the role of Mycoplasma ovipneumoniae. N Z Vet J. 1999;47:155–60. DOIPubMedGoogle Scholar
- Subramaniam R, Herndon CN, Shanthalingam S, Dassanayake RP, Bavananthasivam J, Potter KA, Defective bacterial clearance is responsible for the enhanced lung pathology characteristic of Mannheimia haemolytica pneumonia in bighorn sheep. Vet Microbiol. 2011;153:332–8. DOIPubMedGoogle Scholar
- Cassirer EF, Oldenburg LE, Coggins V, Fowler P, Rudolph KM, Hunter DL, Overview and preliminary analysis of Hells Canyon bighorn sheep die-off, 1995–6. Proceedings of the Tenth Biennial Symposium of the Northern Wild Sheep and Goat Council. 1996 Apr 29–May 3; Silverthorne, Colorado. Cody (WY): The Council; 1996;10:78–86.
Figure
Tables
Cite This ArticleTable of Contents – Volume 18, Number 3—March 2012
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Thomas E. Besser, Washington Animal Disease Diagnostic Laboratory, PO Box 647034, Washington State University, Pullman, WA 99164-7034, USA
Top