Characterization of Virulent West Nile Virus Kunjin Strain, Australia, 2011
Melinda J. Frost
1, Jing Zhang
1, Judith H. Edmonds
1, Natalie A. Prow
1, Xingnian Gu, Rodney Davis, Christine Hornitzky, Kathleen E. Arzey, Deborah Finlaison, Paul Hick, Andrew Read, Jody Hobson-Peters, Fiona J. May, Stephen L. Doggett, John Haniotis, Richard C. Russell, Roy A. Hall
2, Alexander A. Khromykh
2, and Peter D. Kirkland
2
Author affiliations: Elizabeth Macarthur Agriculture Institute, Menangle, New South Wales, Australia (M.J. Frost, J. Zhang, X. Gu, R. Davis, C. Hornitzky, K.E. Arzey, D. Finlaison, P. Hick, A. Read, P.D. Kirkland); The University of Queensland, St Lucia, Queensland, Australia (J.H. Edmonds, N.A Prow, J. Hobson-Peters, F.J. May, R.A. Hall, A. A. Khromykh); University of Sydney and Westmead Hospital, Westmead, New South Wales, Australia (S.L. Doggett, J. Haniotis, R.C. Russell)
Main Article
Figure 2
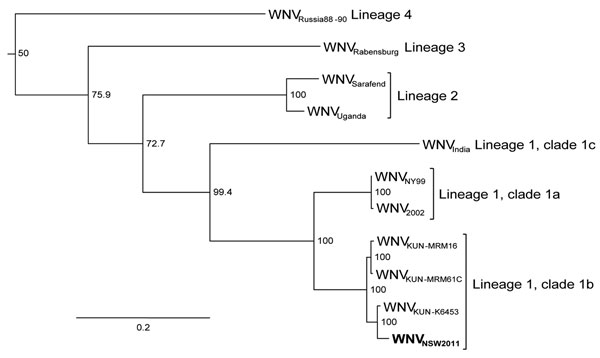
Figure 2. . . Maximum-likelihood tree based on nucleotide sequences of the complete open reading frame of genomes of West Nile virus (WNV) NSW2011 (boldface) and representative strains of WNV from the different lineages and clades. All published complete Kunjin (KUN) virus sequences are included. Bootstrap values are shown on the nodes and are expressed as a percentage of 1,000 replicates. Sequences downloaded from GenBank were WNVRussia88–90, AY277251; WNVRabensburg, AY765264; WNVSarafend, AY688948; WNVUganda, AY532665; WNVIndia, DQ256376; WNVNY99, AF196835; WNV2002, GU827998; WNVKUNV-MRM16, GQ851602; WNVKUNV-MRM61C, AY274504; and WNVKUNV-K6453, GQ851603. NY, New York; NSW, New South Wales. Horizontal branch lengths indicate genetic distance proportional to the scale bar.
Main Article
Page created: April 12, 2012
Page updated: April 12, 2012
Page reviewed: April 12, 2012
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
