Volume 19, Number 12—December 2013
Research
Zoonotic Chlamydiaceae Species Associated with Trachoma, Nepal
Figure 2
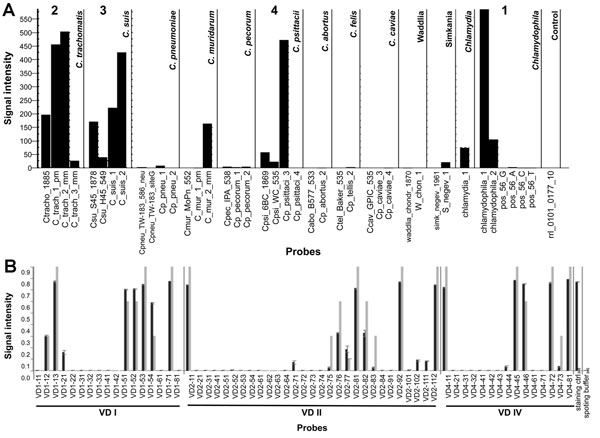
Figure 2. . . Identification of Chlamydiaceae triple infection by using the ArrayTube (Alere Technologies, Jena, Germany) assay. A) Biotinylated PCR product from a DNA extract was hybridized to a DNA microarray carrying species-specific probes from the 23S rRNA gene locus (17). Bar graph shows specific hybridization signals for genus Chlamydia (1), C. trachomatis (2), C. suis (3), and C. psittaci (4) in sample 67. Other signals represent nonspecific cross-hybridization. B) ompA genotyping of the C. trachomatis strain from sample 64 conducted by using the ArrayStrip platform that is specific for C. trachomatis. The best match of this sample was genotype C. The genotype has been determined by automatic comparison of experimentally obtained (black bars) and theoretically constructed (gray bars) hybridization patterns with use of the software's PatternMatch algorithm. The numerical values of matching score MS (measure of similarity between sample and reference strain) and Delta MS (numerical difference between best and second best match) indicate that the identification is highly accurate (21). The rightmost bars represent internal staining control (biotinylated oligonucleotide probe) and spotting buffer (background).
References
- Dean D. Pathogenesis of chlamydial ocular infections. In: Tasman W, Jaeger EA, editors. Duane's foundations of clinical ophthalmology. Philadelphia: Lippincott Williams & Wilkins; 2010. p. 678–702.
- Zhang H, Kandel RP, Sharma B, Dean D. Risk factors for recurrence of postoperative trichiasis: implications for trachoma blindness prevention. Arch Ophthalmol. 2004;122:511–6. DOIPubMedGoogle Scholar
- Zhang H, Kandel RP, Atakari HK, Dean D. Impact of oral azithromycin on recurrence of trachomatous trichiasis in Nepal over 1 year. Br J Ophthalmol. 2006;90:943–8. DOIPubMedGoogle Scholar
- Atik B, Thanh TT, Luong VQ, Lagree S, Dean D. Impact of annual targeted treatment on infectious trachoma and susceptibility to reinfection. JAMA. 2006;296:1488–97. DOIPubMedGoogle Scholar
- Chidambaram JD, Alemayehu W, Melese M, Lakew T, Yi E, House J, Effect of a single mass antibiotic distribution on the prevalence of infectious trachoma. JAMA. 2006;295:1142–6. DOIPubMedGoogle Scholar
- Dean D, Kandel RP, Adhikari HK, Hessel T. Multiple Chlamydiaceae species in trachoma: implications for disease pathogenesis and control. PLoS Med. 2008;5:e14. DOIPubMedGoogle Scholar
- Goldschmidt P, Rostane H, Sow M, Goepogui A, Batellier L, Chaumeil C. Detection by broad-range real-time PCR assay of Chlamydia species infecting human and animals. Br J Ophthalmol. 2006;90:1425–9. DOIPubMedGoogle Scholar
- Lietman T, Brooks D, Moncada J, Schachter J, Dawson C, Dean D. Chronic follicular conjunctivitis associated with Chlamydia psittaci or Chlamydia pneumoniae. Clin Infect Dis. 1998;26:1335–40. DOIPubMedGoogle Scholar
- Gaydos CA. Nucleic acid amplification tests for gonorrhea and Chlamydia: practice and applications. Infect Dis Clin North Am. 2005;19:367–86. DOIPubMedGoogle Scholar
- Sachse K, Vretou E, Livingstone M, Borel N, Pospischil A, Longbottom D. Recent developments in the laboratory diagnosis of chlamydial infections. Vet Microbiol. 2009;135:2–21. DOIPubMedGoogle Scholar
- Dean D, Patton M, Stephens RS. Direct sequence evaluation of the major outer membrane protein gene variant regions of Chlamydia trachomatis subtypes D′, I′, and L2′. Infect Immun. 1991;59:1579–82 .PubMedGoogle Scholar
- Geens T, Desplanques A, Van Loock M, Bonner BM, Kaleta EF, Magnino S, Sequencing of the Chlamydophila psittaci ompA gene reveals a new genotype, E/B, and the need for a rapid discriminatory genotyping method. J Clin Microbiol. 2005;43:2456–61. DOIPubMedGoogle Scholar
- Dean D, Bruno WJ, Wan R, Gomes JP, Devignot S, Mehari T, Predicting phenotype and emerging strains among Chlamydia trachomatis infections. Emerg Infect Dis. 2009;15:1385–94. DOIPubMedGoogle Scholar
- Pannekoek Y, Dickx V, Beeckman DS, Jolley KA, Keijzers WC, Vretou E, Multi locus sequence typing of Chlamydia reveals an association between Chlamydia psittaci genotypes and host species. PLoS ONE. 2010;5:e14179. DOIPubMedGoogle Scholar
- Pantchev A, Sting R, Bauerfeind R, Tyczka J, Sachse K. Detection of all Chlamydophila and Chlamydia spp. of veterinary interest using species-specific real-time PCR assays. Comp Immunol Microbiol Infect Dis. 2010;33:473–84. DOIPubMedGoogle Scholar
- Bom RJ, Christerson L, Schim van der Loeff MF, Coutinho RA, Herrmann B, Bruisten SM. Evaluation of high-resolution typing methods for Chlamydia trachomatis in samples from heterosexual couples. J Clin Microbiol. 2011;49:2844–53. DOIPubMedGoogle Scholar
- Sachse K, Hotzel H, Slickers P, Ellinger T, Ehricht R. DNA microarray-based detection and identification of Chlamydia and Chlamydophila spp. Mol Cell Probes. 2005;19:41–50 and. DOIPubMedGoogle Scholar
- Sachse K, Kuehlewind S, Ruettger A, Schubert E, Rohde G. More than classical Chlamydia psittaci in urban pigeons. Vet Microbiol. 2012;157:476–80. DOIPubMedGoogle Scholar
- Borel N, Kempf E, Hotzel H, Schubert E, Torgerson P, Slickers P, Direct identification of chlamydiae from clinical samples using a DNA microarray assay: a validation study. Mol Cell Probes. 2008;22:55–64. DOIPubMedGoogle Scholar
- Gomes JP, Hsia RC, Mead S, Borrego MJ, Dean D. Immunoreactivity and differential developmental expression of known and putative Chlamydia trachomatis membrane proteins for biologically variant serovars representing distinct disease groups. Microbes Infect. 2005;7:410–20. DOIPubMedGoogle Scholar
- Ruettger A, Feige J, Slickers P, Schubert E, Morre SA, Pannekoek Y, Genotyping of Chlamydia trachomatis strains from culture and clinical samples using an ompA-based DNA microarray assay. Mol Cell Probes. 2011;25:19–27. DOIPubMedGoogle Scholar
- Kern DG, Neill MA, Schachter J. A seroepidemiologic study of Chlamydia pneumoniae in Rhode Island. Evidence of serologic cross-reactivity. Chest. 1993;104:208–13. DOIPubMedGoogle Scholar
- Mahmoud E, Elshibly S, Mardh PA. Seroepidemiologic study of Chlamydia pneumoniae and other chlamydial species in a hyperendemic area for trachoma in the Sudan. Am J Trop Med Hyg. 1994;51:489–94 .PubMedGoogle Scholar
- Lienard J, Croxatto A, Aeby S, Jaton K, Posfay-Barbe K, Gervaix A, Development of a new Chlamydiales-specific real-time PCR and its application to respiratory clinical samples. J Clin Microbiol. 2011;49:2637–42 and. DOIPubMedGoogle Scholar
- Blumer S, Greub G, Waldvogel A, Hassig M, Thoma R, Tschuor A, Waddlia, Parachlamydia and Chlamydiaceae in bovine abortion. Vet Microbiol. 2011;152:385–93. DOIPubMedGoogle Scholar
- Polkinghorne A, Borel N, Becker A, Lu ZH, Zimmermann DR, Brugnera E, Molecular evidence for chlamydial infections in the eyes of sheep. Vet Microbiol. 2009;135:142–6. DOIPubMedGoogle Scholar
- Girjes AA, Hugall A, Graham DM, McCaul TF, Lavin MF. Comparison of type I and type II Chlamydia psittaci strains infecting koalas (Phascolarctos cinereus). Vet Microbiol. 1993;37:65–83. DOIPubMedGoogle Scholar
- Becker A, Lutz-Wohlgroth L, Brugnera E, Lu ZH, Zimmermann DR, Grimm F, Intensively kept pigs pre-disposed to chlamydial associated conjunctivitis. J Vet Med A Physiol Pathol Clin Med. 2007;54:307–13. DOIPubMedGoogle Scholar
- Osman KM, Ali HA, Eljakee JA, Galal HM. Prevalence of Chlamydophila psittaci infections in the eyes of cattle, buffaloes, sheep and goats in contact with a human population. Transbound Emerg Dis. 2013;60:245–51 .DOIPubMedGoogle Scholar
- Yamazaki T, Nakada H, Sakurai N, Kuo CC, Wang SP, Grayston JT. Transmission of Chlamydia pneumoniae in young children in a Japanese family. J Infect Dis. 1990;162:1390–2. DOIPubMedGoogle Scholar
- Hughes C, Maharg P, Rosario P, Herrell M, Bratt D, Salgado J, Possible nosocomial transmission of psittacosis. Infect Control Hosp Epidemiol. 1997;18:165–8. DOIPubMedGoogle Scholar
- Lenzko H, Moog U, Henning K, Lederbach R, Diller R, Menge C, High frequency of chlamydial co-infections in clinically healthy sheep flocks. BMC Vet Res. 2011;7:29. DOIPubMedGoogle Scholar
- Harrison HR, Boyce WT, Wang SP, Gibb GN, Cox JE, Alexander ER. Infection with Chlamydia trachomatis immunotype J associated with trachoma in children in an area previously endemic for trachoma. J Infect Dis. 1985;151:1034–6. DOIPubMedGoogle Scholar
- Mordhorst CH, Wang SP, Grayston JT. Childhood trachoma in a nonendemic area. Danish trachoma patients and their close contacts, 1963 to 1973. JAMA. 1978;239:1765–71. DOIPubMedGoogle Scholar
- Forsey T, Darougar S. Acute conjunctivitis caused by an atypical chlamydial strain: Chlamydia IOL 207. Br J Ophthalmol. 1984;68:409–11. DOIPubMedGoogle Scholar
- Wang S, Grayston JT. Trachoma in the Taiwan monkey Macaca cyclopis. Ann N Y Acad Sci. 1962;98:177–87. DOIPubMedGoogle Scholar
- Taylor HR, Prendergast RA, Dawson CR, Schachter J, Silverstein AM. An animal model for cicatrizing trachoma. Invest Ophthalmol Vis Sci. 1981;21:422–33 .PubMedGoogle Scholar
- Collier LH. Experimental infection of baboons with inclusion blennorrhea and trachoma. Ann N Y Acad Sci. 1962;98:188–96. DOIPubMedGoogle Scholar
- Watkins NG, Hadlow WJ, Moos AB, Caldwell HD. Ocular delayed hypersensitivity: a pathogenetic mechanism of chlamydial-conjunctivitis in guinea pigs. Proc Natl Acad Sci U S A. 1986;83:7480–4. DOIPubMedGoogle Scholar