Volume 19, Number 7—July 2013
Dispatch
Undetected Multidrug-Resistant Tuberculosis Amplified by First-line Therapy in Mixed Infection
Abstract
Infections with >1 Mycobacterium tuberculosis strain(s) are underrecognized. We show, in vitro and in vivo, how first-line treatment conferred a competitive growth advantage to amplify a multidrug-resistant M. tuberculosis strain in a patient with mixed infection. Diagnostic techniques that identify mixed tubercle bacilli populations are needed to curb the spread of multidrug resistance.
As the number of multidrug-resistant tuberculosis (TB) cases continues to rise, so does the amplification of multidrug-resistant Mycobacterium tuberculosis strains during treatment (1,2). This amplification is generally assumed to result from in vivo evolution of drug resistance caused by poor therapy compliance or, in high-incidence settings, from exogenous reinfection with a multidrug-resistant strain. We report a case in which emergence of multidrug resistance did not result from in vivo acquisition of drug resistance by a drug-sensitive strain or from exogenous reinfection with an already resistant strain. By integrating epidemiologic, microbiological, and molecular strain typing data with in vitro competitive growth experiments, we provide evidence for an initial mixed infection with a drug-sensitive strain and an undetected drug-resistant strain that outgrew the sensitive strain under the selection pressure of first-line chemotherapy.
M. tuberculosis strains in sputum from TB-infected patients or in samples from the disease site are generally identified by strain typing a single broth culture or colony grown on solid medium. However, this method does not enable identification of mixed infections, and any treatment regimen would be determined on the basis of the drug sensitivity of the strain with the fastest growth rate in the in vitro culture. Use of suboptimal drug combinations could lead to selection of a slower growing, drug-resistant strain already present in the host and thus to treatment failure.
Studies of artificially mixed M. tuberculosis strains before and after culture showed that culturing can reduce the clonal complexity of the strains and that, in most samples (6/10), only 1 strain will be identified in mixed infections after culture (3). This suggests that mixed infections and clonal complexity are underrepresented in culture-based diagnoses of TB. In support of this suggestion, the results of molecular-based methods that use strain-specific PCR showed that 2.1%–19.0% of patients with active TB in moderate to high incidence countries were simultaneously infected with >2 strains (1,2,4–10).
Possible co-infection of patients with drug-sensitive and drug-resistant M. tuberculosis strains has been described (1,2), and modeling of the effect of such co-infection on the long-term dynamics of tuberculous infection has led to the hypothesis that persons with this type of infection may retain small populations of drug-resistant bacteria that can flourish after the host receives treatment (11). van Rie et al. showed the amplification of a drug-resistant strain after treatment and postulated selection of drug-resistant strains from an initial mixed infection through antimicrobial drug pressure (2). We confirm this hypothesis by combining detailed longitudinal clinical and microbiological observation with the use of novel in vitro growth competition assays to study 2 co-infecting patient strains in the presence and absence of the primary drug used in treatment.
The 2 M. tuberculosis strains were isolated from a 68-year-old man from Portugal. He did not have HIV and was treated as a confined inpatient, limiting the possibility that this was not a true in vivo mixed infection. Using a novel approach, we correlated in vitro growth and treatment characteristics for the patient strains with the in vivo strain predominance and persistence of a less-fit, drug-resistant strain. All samples were obtained with approval from St. Mary Hospital’s (London, UK) Research Ethics Center (no. 07/H0712/85) and with the patient’s written informed consent.
Details of the patient samples are in the Table. The initial bronchoalveolar lavage smear sample was positive for acid-fast bacilli (AFB); culture results were positive for fully sensitive M. tuberculosis. Treatment with isoniazid, rifampin, ethambutol, and pyrazinamide was begun. Because of the patient’s alcohol use, his treatment was managed on an inpatient basis in a single-patient, negative-pressure room. Two months later, repeat sputum smears were positive for AFB, and culture results were positive for fully sensitive M. tuberculosis. After 4 months of treatment, the patient’s clinical signs had not improved, and his sputum smear was still positive for AFB. Culture results for the sputum sample were positive for M. tuberculosis resistant to isoniazid and ethambutol; a modified treatment regime resolved the infection, and the patient was released the following month, by which time his smear and culture results were negative.
We molecularly characterized the strains by using mycobacterial interspersed repetitive unit–variable number tandem repeat (MIRU-VNTR) typing (12); results showed that the drug-sensitive and drug-resistant M. tuberculosis strains were 2 distinct strains (Table) rather than 1 sensitive strain that had become resistant through mutagenesis. Because the patient was isolated while an inpatient, exogenous reinfection with a primary drug-resistant strain was ruled out. In addition, treatment compliance was directly observed, so in vivo development of drug resistance caused by poor compliance was also ruled out. Thus, it is highly likely that the patient was harboring a mixed infection of drug-sensitive and drug-resistant strains when he initially sought care at the clinic. Such a co-infection would not have been detected because single-colony or broth cultures are commonly used for strain typing, and these techniques would give the fastest growing strain a competitive advantage. Thus, we devised a competitive growth assay to determine if the patient had a mixed infection and to provide correlating in vitro and in vivo evidence of mixed infection (Figure 1).
For the in vitro growth analyses of the 2 strains, we inoculated broth cultures and measured growth at an optical density of 600 nm, characterized the dominant strain by using MIRU-VNTR, and quantified colony-forming units on agar plates in the presence and absence of isoniazid. At several points during logarithmic growth, the drug-sensitive strain grew substantially faster than the resistant strain (Figure 2, panel A), suggesting that without the selective pressure of isoniazid, the sensitive strain would be most prevalent in a mixed infection.
For the in vitro competition assays, the strains were mixed (1:1), and isoniazid (0.2 μg/mL) was or was not added before measurement of growth and determination of the dominant strain. In the presence of isoniazid, the growth rate was lower, suggesting that the drug-resistant strain outcompeted the drug-sensitive strain to become the dominant strain (Figure 2, panel B). This was confirmed by MIRU-VNTR typing and growth analyses (Figure 2, panel C). These results indicate that 1) the drug-sensitive strain had a competitive growth advantage, causing this strain type to be identified as the sole infecting strain, and that 2) the drug-resistant strain gained the competitive advantage when isoniazid was added and became the predominant strain after treatment. These findings correlate precisely with the patient data (Table) and, we believe, is representative of the in vivo host infection.
We show the selection and subsequent clinically relevant emergence of a drug-resistant M. tuberculosis strain after treatment of a drug-sensitive strain in a patient with an initial mixed infection. This case illustrates the prospect of treatment failure for TB caused by mixed infection with strains with different drug susceptibility and growth rates. The proportion of cases of secondary multidrug resistance caused by such initial mixed infections is not known; however, the ability of the resistant strain to outcompete the sensitive strain under treatment and then to potentially transmit further may have substantial implications for the control and prevention of multidrug resistance.
The case also highlights the urgent need for improved diagnostic techniques that can routinely identify mixed populations of tubercle bacilli. Given the difficulty of detecting TB co-infections by using routine diagnostic microbiology techniques, co-infection is likely underrecognized. Co-infection can currently be ruled out only by using specialized techniques, such as molecular analysis of original sample (pre-culture); analysis of multiple colonies; or the GeneXpert assay (Cepheid, Sunnyvale, CA, USA) (13). Rapid detection of mixed infections with distinct drug susceptibility profiles would enable suitably tailored drug regimens from the start of treatment, which could prevent treatment failure and emergence and transmission of drug-resistant strains.
Dr Hingley-Wilson is a senior postdoctoral scientist in the Faculty of Health and Medical Sciences at the University of Surrey, Guildford, UK. Her primary research interest is the host–pathogen interaction in tuberculosis.
Acknowledgments
We thank the patient and the staff of St Mary’s Tuberculosis Service. We also thank Saranya Sridhar for his help with the statistical analyses and Kathryn Lougheed and Dany Beste for help with manuscript preparation.
Financial support was provided by the Wellcome Trust. A.L. is a Wellcome Senior Clinical Research Fellow and National Institute of Health Research Senior Investigator.
The experiments were designed and conducted by S.M. H-W., and the paper was written by S.M. H-W, R.C. and A.L.
References
- Mendez MP, Landon ME, McCloud MK, Davidson P, Christensen PJ. Co-infection with pansensitive and multidrug-resistant strains of Mycobacterium tuberculosis. Emerg Infect Dis. 2009;15:578–80 . DOIPubMedGoogle Scholar
- van Rie A, Victor TC, Richardson M, Johnson R, van der Spuy GD, Murray EJ, Reinfection and mixed infection cause changing Mycobacterium tuberculosis drug-resistance patterns. Am J Respir Crit Care Med. 2005;172:636–42 . DOIPubMedGoogle Scholar
- Martín A, Herranz M, Ruiz Serrano MJ, Bouza E, Garcia de Viedma D. The clonal composition of Mycobacterium tuberculosis in clinical specimens could be modified by culture. Tuberculosis (Edinb). 2010;90:201–7. DOIPubMedGoogle Scholar
- Warren RM, Victor TC, Streicher EM, Richardson M, Beyers N, Gey van Pittius NC, Patients with active tuberculosis often have different strains in the same sputum specimen. Am J Respir Crit Care Med. 2004;169:610–4 . DOIPubMedGoogle Scholar
- Wang JY, Hsu HL, Yu MC, Chiang CY, Yu FL, Yu CJ, Mixed infection with Beijing and non-Beijing strains in pulmonary tuberculosis in Taiwan: prevalence, risk factors, and dominant strain. Clin Microbiol Infect. 2011;17:1239–45. DOIPubMedGoogle Scholar
- Huang HY, Tsai YS, Lee JJ, Chiang MC, Chen YH, Chiang CY, Mixed infection with Beijing and non-Beijing strains and drug resistance pattern of Mycobacterium tuberculosis. J Clin Microbiol. 2010;48:4474–80. DOIPubMedGoogle Scholar
- Mallard K, McNerney R, Crampin AC, Houben R, Ndlovu R, Munthali L, Molecular detection of mixed infections of Mycobacterium tuberculosis strains in sputum samples from patients in Karonga District, Malawi. J Clin Microbiol. 2010;48:4512–8. DOIPubMedGoogle Scholar
- Shamputa IC, Rigouts L, Eyongeta LA, El Aila NA, van Deun A, Salim AH, Genotypic and phenotypic heterogeneity among Mycobacterium tuberculosis isolates from pulmonary tuberculosis patients. J Clin Microbiol. 2004;42:5528–36. DOIPubMedGoogle Scholar
- Navarro Y, Herranz M, Pérez-Lago L, Martinez Lirola M, Ruiz-Serrano MJ, Bouza E, Systematic survey of clonal complexity in tuberculosis at a populational level and detailed characterization of the isolates involved. J Clin Microbiol. 2011;49:4131–7. DOIPubMedGoogle Scholar
- Braden CR, Morlock GP, Woodley CL, Johnson KR, Colombel AC, Cave MD, Simultaneous infection with multiple strains of Mycobacterium tuberculosis. Clin Infect Dis. 2001;33:e42–7. DOIPubMedGoogle Scholar
- Sergeev R, Colijn C, Cohen T. Models to understand the population-level impact of mixed strain M. tuberculosis infections. J Theor Biol. 2011;280:88–100. DOIPubMedGoogle Scholar
- Evans JT, Hawkey PM, Smith EG, Boese KA, Warren RE, Hong G. Automated high-throughput mycobacterial interspersed repetitive unit typing of Mycobacterium tuberculosis strains by a combination of PCR and nondenaturing high-performance liquid chromatography. J Clin Microbiol. 2004;42:4175–80. DOIPubMedGoogle Scholar
- Ioannidis P, Papventsis D, Karabela S, Nikolaou S, Panagi M, Raftopoulou E, Cepheid GeneXpert MTB/RIF assay for Mycobacterium tuberculosis detection and rifampin resistance identification in patients with substantial clinical indication of tuberculosis and smear-negative microscopy results. J Clin Microbiol. 2011;49:3068–70. DOIPubMedGoogle Scholar
Figures
Table
Cite This ArticleTable of Contents – Volume 19, Number 7—July 2013
| EID Search Options |
|---|
|
|
|
|
|
|
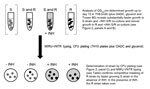
![Thumbnail of Analyses of the amplification of multidrug-resistant Mycobacterium tuberculosis during treatment of a drug-sensitive (S) strain in a mixed infection (i.e., infection with drug-resistant [R] and S strains). In the presence of isoniazid (INH), the faster growing S strain lost its competitive advantage, and the R strain became more prevalent. A–C) Data are means of 3 independent replicates with SE bars. A) Single strain growth analyses of S (black circles) and R (gray triangles) M. tub](/eid/images/13-0313-F2-tn.jpg)
Please use the form below to submit correspondence to the authors or contact them at the following address:
Suzanne M. Hingley-Wilson, Department of Microbial and Cellular Sciences, University of Surrey, Stag Hill Campus, Guildford, Surrey, GU3 1DY, UK
Top