Volume 21, Number 12—December 2015
Research
High Prevalence of Intermediate Leptospira spp. DNA in Febrile Humans from Urban and Rural Ecuador
Figure
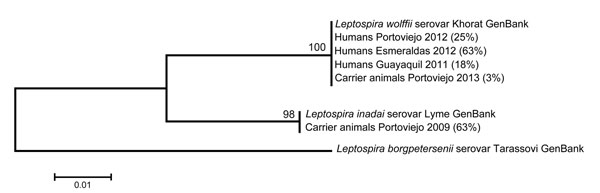
Figure. Maximum-likelihood tree for DNA sequences of the Leptospira spp. rrs gene recovered from serum samples from febrile humans and from urine and kidney samples from animal carriers in Ecuador. Esmeraldas, Portoviejo, and Guayaquil are 3 rural, semiurban, and urban communities, respectively, along the coast of Ecuador. Pathogenic L. borgpetersenii was used as an outgroup. Numbers in parentheses indicate the percentage of samples per community that contained DNA signatures highly similar to GenBank reference strains L. wolffii (NR_044042), L. inadai (accession no. JQ988844.1), and L. borgpetersenii (accession no. JQ988861.1). Scale bar indicates the degree of nucleotide substitutions.
Page created: November 16, 2015
Page updated: November 16, 2015
Page reviewed: November 16, 2015
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.