Volume 21, Number 7—July 2015
Dispatch
Novel Arenavirus Isolates from Namaqua Rock Mice, Namibia, Southern Africa
Abstract
Arenaviruses are feared as agents that cause viral hemorrhagic fevers. We report the identification, isolation, and genetic characterization of 2 novel arenaviruses from Namaqua rock mice in Namibia. These findings extend knowledge of the distribution and diversity of arenaviruses in Africa.
Arenaviruses are known to cause severe hemorrhagic fevers across the globe with case fatality rates up to 30% (1). The viruses possess a bisegmented, single-stranded RNA genome with ambisense coding strategy consisting of a small segment coding for the nucleoprotein and glycoprotein and a large (L) segment coding for the RNA-dependent RNA polymerase and matrix protein.
In Africa, Lassa virus (LASV) and Lujo virus are the only known members of the family Arenaviridae that cause human disease (2,3); however, evidence for lymphocytic choriomeningitis virus, another Arenaviridae sp., was recently reported in Gabon (4). Several other arenaviruses of unknown pathogenic potential have also been found in Africa: Gbagroube, Kodoko, and Menekre viruses from western Africa (5,6); Ippy (IPPYV) and Mobala viruses from central Africa; Mopeia, Morogoro, Luna, and Lunk viruses from eastern Africa; and Merino Walk virus (MWV) from southern Africa (7,8). All of these viruses are carried by rodents of the family Muridae.
Until now, no molecular detection of arenaviruses has been reported from Namibia. A study in 1991 described a low seroprevalence (0.8%) for LASV antibodies in humans in northern Namibia (9). Because of lack of data about arenavirus occurrence and effects in southwestern Africa, we conducted a study of small mammals from Namibia to detect infection with arenaviruses.
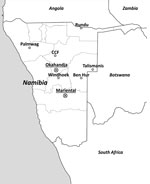
During 2010–2012, animal trapping was performed in 8 areas in central and northern Namibia (Figure 1), and samples from 812 rodents and shrews were obtained (Table 1). The animals were dissected in the field and stored individually in a field freezer at −20°C and later at −80°C. For primary arenavirus screening, lung sections of all animals were homogenized, and RNA was extracted and reversely transcribed by using random hexamer primers. Screening was performed by arenavirus genus-specific reverse transcription PCR (RT-PCR) (10) to detect the L genomic segment. From samples testing positive by arenavirus PCR, frozen lung tissue aliquots were homogenized and added to confluent Vero-E6 cells (ATCC CRL-1586; American Type Culture Collection, Manassas, VA, USA) for virus isolation.
For genome sequencing, pellets from ultracentrifuged supernatant of infected cell cultures were lysed, and total RNA was purified. RNA was then subjected to random-primed RT-PCR as described (11). Next-generation sequencing was performed by using a 454 Genome Sequencer Junior (Roche, Indianapolis, IN, USA), and results were aligned against the virus database by using blastn and blastx algorithms (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequencing results matching arenavirus sequences were mapped to the LASV strain Josiah (GenBank accession no. AY628203). Because of low coverage for N27, an additional MiSeq (Illumina, San Diego, CA, USA) run was performed. De novo assembly of the data was performed with Geneious software (Biomatters, Auckland, New Zealand) (12). Sequence gaps or regions with low coverage were verified by Sanger sequencing (Applied Biosystems, Foster City, CA, USA). Genome segment outermost noncoding termini were sequenced after linkage by T4-RNA-Ligase (New England Biolabs, Ipswich, MA, USA) and RT-PCR amplification.
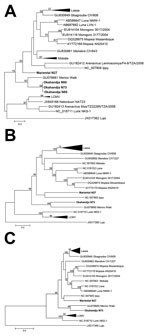
Of the 812 rodents and shrews tested (Table 1), arenavirus RNA was found in lung tissue samples of 4 Namaqua rock mice (Micaelamys [Aethomys] namaquensis), 3 from Okahandja (N73, N80, N85) and 1 from Mariental (N27). Sanger sequencing of PCR products from a 338-nt fragment of the viral polymerase gene confirmed arenavirus-like origin. Initial phylogenetic analysis showed that the Okahandja specimens were related to MWV, but the sample from Mariental was a highly divergent member of the genus Arenavirus (Figure 2, panel A). Cell culture isolation was performed with samples N27 and N73 and resulted in 2 novel arenavirus isolates: Mariental virus (MRTV) and Okahandja virus (OKAV), respectively.
The genomes of the 2 arenaviruses were investigated by using next-generation sequencing and RT-PCR Sanger sequencing. The genome data obtained for MRTV and OKAV showed a typical arenavirus nucleotide composition with the L segment (MRTV: 6,840 nt; OKAV: 7,170 nt) coding for RNA polymerase and matrix protein and the S segment (MRTV: 3,360 nt; OKAV: 3,379 nt) coding for glycoprotein and nucleocapsid protein. Table 2 shows the nucleotide and amino acid sequence identities of nucleocapsid and glycoprotein open reading frame with other Old World (i.e., Eastern Hemisphere locations such as Europe, Asia, Africa) representatives of genus Arenavirus. On the basis of the nucleocapsid amino acid identity, OKAV is most related to MWV (75.7% identity). Furthermore, MRTV has the highest amino acid identity with IPPYV (71.4% identity) and with Gbagroube, Lassa, Luna, and Mobala viruses (≈70% identity).
In the nucleocapsid-based phylogenetic tree, OKAV clusters with 100% bootstrap support with MWV detected in Myotomys unisulcatus rodents in South Africa (Figure 2, panel B), and MRTV forms a clade with IPPYV isolated from Praomys spp. in the Central African Republic. The bootstrap support of this monophyletic group of the tree lies at 56%. The analysis of the glycoprotein open reading frame (Figure 2, panel C) leads to a similar result; OKAV shares the most recent common ancestor with MWV, and MRTV clusters with IPPYV but with a weaker bootstrap support.
We detected and isolated 2 novel arenaviruses in Namibia, OKAV and MRTV. OKAV clearly clustered in relationship with the MWV from southern Africa, but MRTV is a more divergent member of the Old World arenavirus clade. According to amino acid identity and phylogenetic analysis, MRTV was most closely related to IPPYV from the Central African Republic; however, the low bootstrap support precluded a stringent estimation of this closest relative.
These new strains comply with the arenavirus species definition (14) on the basis of amino acid differences in nucleocapsid of >12% (>20% for both viruses), presence of specific host species, and existence of laboratory isolates. These properties indicate that MRTV and OKAV represent distinct arenavirus species.
These 2 viruses were found in the same host species located within a radius of 300 km. MRTV was found in only 1 sample (of 266); OKAV was detected in samples from 3 animals. Although more unlikely for OKAV than for MRTV, the possibility of a spillover infection to M. namaquensis from a still unknown reservoir host cannot be ruled out for either virus.
The Namaqua rock mouse’s habitat includes the tree and shrub savannahs of Namibia and most parts of southern Africa, including Namibia, South Africa, Botswana, Zimbabwe, and parts of Mozambique (15). These locations imply the possible occurrence of MRTV or OKAV in larger regions of the continent. Cell culture isolates and genomic sequence data are the first prerequisites for evaluating the public health relevance of these new viruses. Our findings extend the knowledge of geographic distribution and genetic diversity of arenaviruses in Africa.
Dr. Witkowski is a postdoctoral researcher at the Institute of Virology of the Charité Medical School in Berlin, Germany. His research interests are viral emerging infectious diseases on the African continent and their clinical impact and evolution.
Acknowledgments
We thank C. Chimimba for advice in small mammal systematics, P. Chimwamurombe for advice during preliminary screening for arenaviruses, and C. Priemer for technical support.
Trapping in Namibia was carried out under research permit nos. 1572/2011, 1666/2012 and 1794/2013, granted by Namibia’s Ministry of Environment and Tourism.
This study was supported by Deutsche Forschungsgemeinschaft (grant KR1293/13-1) and by the Slovak Research and Development Agency under the contract no. DO7RP-0008-09.
References
- Briese T, Paweska JT, McMullan LK, Hutchison SK, Street C, Palacios G, Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 2009;5:e1000455. DOIPubMedGoogle Scholar
- Frame JD, Baldwin JM, Gocke DJ, Troupt AM. Lassa fever, a new virus disease of man from West Africa. Am J Trop Med Hyg. 1970;19:670–6.PubMedGoogle Scholar
- N’Dilimabaka N, Berthet N, Rougeron V, Mangombi JB, Durand P, Maganga GD, Evidence of lymphocytic choriomeningitis virus (LCMV) in domestic mice in Gabon: risk of emergence of LCMV encephalitis in Central Africa. J Virol. 2015;89:1456–60. DOIPubMedGoogle Scholar
- Lecompte E, terMeulen J, Emonet S, Daffis S, Charrel RN. Genetic identification of Kodoko virus, a novel arenavirus of the African pigmy mouse (Mus (Nannomys) minutoides) in West Africa. Virology. 2007;364:178–83. DOIPubMedGoogle Scholar
- Coulibaly-N’Golo D, Allali B, Kouassi SK, Fichet-Calvet E, Becker-Ziaja B, Rieger T, Novel arenavirus sequences in Hylomyscus sp. and Mus (Nannomys) setulosus from Côte d’Ivoire: implications for evolution of arenaviruses in Africa. PLoS ONE. 2011;6:e20893. DOIPubMedGoogle Scholar
- Ishii A, Thomas Y, Moonga L, Nakamura I, Ohnuma A, Hang’ombe B, Novel arenavirus, Zambia. Emerg Infect Dis. 2011;17:1921–4. DOIPubMedGoogle Scholar
- Zapata JC, Salvato MS. Arenavirus variations due to host-specific adaptation. Viruses. 2013;5:241–78. DOIPubMedGoogle Scholar
- Joubert JJ, van der Merve CA, Lourens JH, Lecatsas G, Siegrühn C. Serological markers of hepatitis B virus and certain other viruses in the population of eastern Caprivi, Namibia. Trans R Soc Trop Med Hyg. 1991;85:101–3. DOIPubMedGoogle Scholar
- Vieth S, Drosten C, Lenz O, Vincent M, Omilabu S, Hass M, RT-PCR assay for detection of Lassa virus and related Old World arenaviruses targeting the L gene. Trans R Soc Trop Med Hyg. 2007;101:1253–64. DOIPubMedGoogle Scholar
- Palacios G, Quan P, Jabado OJ, Conlan S, Hirschberg DL, Liu Y, Panmicrobial oligonucleotide array for diagnosis of infectious diseases. Emerg Infect Dis. 2007;13:73–81. DOIPubMedGoogle Scholar
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–9. DOIPubMedGoogle Scholar
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. DOIPubMedGoogle Scholar
- Index of viruses—arenaviridae. In: Büchen-Osmond C, editor. The Universal Virus Database, version 4. New York: Columbia University, 2006 [cited 2014 Aug 11]. www.ncbi.nlm.nih.gov/ICTVdb/Ictv/fs_index.htm
- Coetzee N, Griffin M, Taylor PJ. Aethomys namaquensis. In: The International Union for Conservation of Nature Red List of Threatened Species. Version 2014.1. 2014 [cited 2014 Aug 11]. www.iucnredlist.org
Figures
Tables
Cite This ArticleTable of Contents – Volume 21, Number 7—July 2015
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Peter T. Witkowski, Institute of Virology, Charité Medical School, Charitéplatz 1, 10117 Berlin, Germany
Top