Volume 4, Number 2—June 1998
Perspective
Wild Primate Populations in Emerging Infectious Disease Research: The Missing Link?
Figure 2
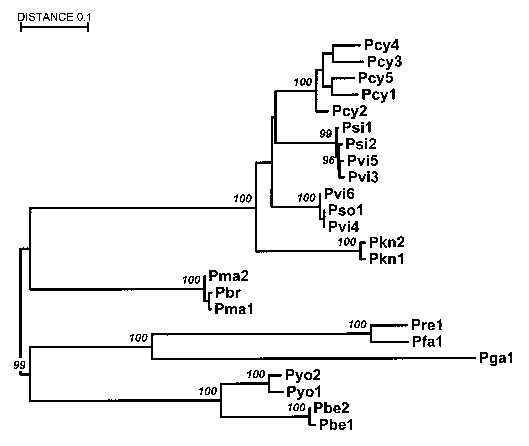
Figure 2. Relationships between primate and human parasites: Malaria phylogeny based on the circumsporozoite protein gene. The alignment does not include the central repeat region. P. falciparum (Pfa), P. vivax (Pvi), and P. malariae (Pma) are from humans; P. cynomolgi (Pcy), P. simiovale (Pso), and P. knowlesi (Pkn) are from macaques; P. simium (Psi) and P. brasilianum (Pbr) are from New World monkeys; P. reichenowi (Pre) is from chimpanzees; P. gallinacium (Pga) is from birds; and P. berghei (Pbe) and P. yoelii (Pyo) are from rodents. The numbers in the names indicate different isolates as described in (30). The sequence of P. gallinacium was reported by (31). The numbers on the branches are bootstrap % based on 500 pseudoreplications. The tree was estimated by the neighbor-joining method with the Tajima and Nei distance (32).
References
- Ruch TC. Diseases of laboratory primates. Philadelphia: W.B. Saunders Company; 1959.
- Brack M. Agents transmissible from simians to man. Berlin: Springer-Verlag; 1987.
- Morse SS. Emerging viruses. In: Morse SS, editor. Emerging viruses. New York: Oxford University Press, Inc.; 1993.
- Levins R, Awerbuch T, Brinkmann U, Eckardt I, Epstein P, Makhoul N, The emergence of new diseases. Am Sci. 1994;82:52–60.
- Wilson ME, Levins R, Spielman A. Disease in evolution: global changes and emergence of infectious diseases. Ann N Y Acad Sci. 1994;:740.
- Mutombo M, Arita I, Jezek Z. Human monkeypox transmitted by a chimpanzee in a tropical rain-forest area of Zaire. Lancet. 1983;34:735–7. DOIGoogle Scholar
- Eidson M, Tierney LA, Rollag OJ, Becker T, Brown T, Hull HF. Feline plague in New Mexico: risk factors and transmission to humans. Am J Public Health. 1988;78:1333–5. DOIPubMedGoogle Scholar
- Douglas JD, Soike KF, Raynor J. The incidence of poliovirus in chimpanzees (Pan troglodytes). Lab Anim Care. 1970;20:265–8.PubMedGoogle Scholar
- Morell V. Chimpanzee outbreak heats up search for Ebola origin. Science. 1995;268:974–5. DOIPubMedGoogle Scholar
- Meyers WM, Gormus BJ, Walsh GP, Baskin B, Hubbard GB. Naturally acquired and experimental leprosy in nonhuman primates. Am J Trop Med Hyg. 1991;44:24–7.PubMedGoogle Scholar
- McCallum H, Dobson A. Detecting disease and parasite threats to endangered species and ecosystems. Trends Ecol Evol. 1995;10:190–4. DOIGoogle Scholar
- Coatney GR, Collins WE, Warren M, Contacos PG. The primate malarias. Bethesda: U.S. Department of Health, Education, and Welfare; 1971.
- Beverley-Burton M, Crichton VF. Attempted experimental cross infections with mammalian guinea-worms, Drancunculus spp. (Nematoda: Dracunculoidea). Am J Trop Med Hyg. 1976;25:704–8.PubMedGoogle Scholar
- Fuller GK, Lemma A, Haile T. Schistosomiasis in Omo National Park of Southwest Ethiopia. Am J Trop Med Hyg. 1979;28:526–30.PubMedGoogle Scholar
- Shah KV. A review of the circumstances and consequences of simian virus SV40 contamination of human vaccines. Symposium on Continuous Cell Lines as Substrates for Biologicals. Developments in biological standardization, Vol. 70; 1989.
- Khabbaz RF, Heneine W, George JR, Parekh B, Rowe T, Woods T, Brief report: infection of a laboratory worker with simian immunodeficiency virus. N Engl J Med. 1994;330:172–7. DOIPubMedGoogle Scholar
- Michler RE. Xenotransplantation: risks, clinical potential, and future prospects. Emerg Infect Dis. 1996;2:64–70. DOIPubMedGoogle Scholar
- Berkelman RL, Pinner RW, Hughes JM. Addressing emerging microbial threats in the United States. JAMA. 1996;275:315–7. DOIPubMedGoogle Scholar
- Lederberg J, Shope RE, Oaks SC. Emerging infections: microbial threats to health in the United States. Washington: National Academy Press; 1992.
- LeDuc JW. World Health Organization strategy for emerging infectious diseases. JAMA. 1996;275:318–20. DOIPubMedGoogle Scholar
- Meslin FX. Surveillance and control of emerging zoonoses. World Health Stat Q. 1992;45:200–7.PubMedGoogle Scholar
- Holmes ED, Nee S, Rambaut A, Garnett GP, Harvey PH. Revealing the history of infectious disease epidemics through phylogenetic trees. Philos Trans R Soc Lond B Biol Sci. 1995;349:33–40. DOIPubMedGoogle Scholar
- Kalter SS, Heberling RL, Cooke AW, Barry JD, Tian PY, Northam WJ. Viral infections of non-human primates. Lab Anim Sci. 1997;47:461–7.PubMedGoogle Scholar
- Myers G, MacInnes K, Korber B. The emergence of simian/human immunodeficiency viruses. AIDS Res Hum Retroviruses. 1992;8:373–86. DOIPubMedGoogle Scholar
- Peeters M, Fransen K, Delaporte E, Van Den Haesevelde M, Gershy-Damet GM, Kestens L, Isolation and characterization of a new chimpanzee lentivirus (simian immunodeficiency virus isolate cpz-ant) from a wild-captured chimpanzee. AIDS. 1992;6:447–51.PubMedGoogle Scholar
- Huet TR, Cheynier A, Meyerhans A, Roelants G, Wain-Hobson S. Genetic organization of a chimpanzee lentivirus related to HIV-1. Nature. 1990;345:356–459. DOIPubMedGoogle Scholar
- Mindell DP. Positive selection and rates of evolution in immunodeficiency viruses from humans and chimpanzees. Proc Natl Acad Sci U S A. 1996;93:3284–8. DOIPubMedGoogle Scholar
- Liu HF, Goubau P, Van Brussel M, Van Laethem K, Chen YC, Desmyter J, The three human T-lymphotropic virus type I subtypes arose from three geographically distinct simian reservoirs. J Gen Virol. 1996;77:359–68. DOIPubMedGoogle Scholar
- Collins WE. Major animal models in malaria research: simian. In: Wernsdorfer WH, editor. Malaria: principles and practice of malariology. Edinburgh: Churchill Livingstone; 1988.
- Escalante AA, Barrio E, Ayala FJ. Evolutionary origin of human and primate malarias: evidence from the circumsporozoite protein gene. Mol Biol Evol. 1995;12:616–26.PubMedGoogle Scholar
- McCutchan TF, Kissinger JC, Touray MG, Rogers MJ, Li J, Sullivan M, Comparison of circumsporozoite proteins from avian and mammalian malaria: biological and phylogenetic implications. Proc Natl Acad Sci U S A. 1996;93:11889–94. DOIPubMedGoogle Scholar
- Tajima F, Nei M. Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol. 1984;1:269–85.PubMedGoogle Scholar
- Qari SH, Shi YP, Pieniazek NJ, Collins WE, Lal AA. Phylogenetic relationship among the malaria parasites based on small subunit rRNA gene sequences: monophyletic nature of the human malaria parasite, Plasmodium falciparum. Mol Phylogenet Evol. 1996;6:157–65. DOIPubMedGoogle Scholar
- Waters AP, Higgins DG, McCutchan TF. Evolutionary relatedness of some primate models of Plasmodium. Mol Biol Evol. 1993;10:914–23.PubMedGoogle Scholar
- Escalante AA, Ayala FJ. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proc Natl Acad Sci U S A. 1994;91:11373–7. DOIPubMedGoogle Scholar
- Qari SH, Shi YP, Povoa MM, Alpers MP, Deloron P, Murphy GS, Global occurrence of Plasmodium vivax-like human malaria parasite. J Infect Dis. 1993;168:1485–9.PubMedGoogle Scholar
- Desrosiers RC. HIV-1 origins: a finger on the missing link. Nature. 1990;345:288–9. DOIPubMedGoogle Scholar
- Peeters M, Janssens W, Fransen K, Brandful J, Heyndrickx L, Koffi K, Isolation of simian immunodeficiency viruses from two sooty mangabeys in Côte d'Ivoire: virological and genetic characterization and relationship to other HIV type 2 and SIVsm/mac strains. AIDS Res Hum Retroviruses. 1994;10:1289–94. DOIPubMedGoogle Scholar
- Brooks DR, McLennan DA. Parascript: parasites and the language of evolution. Washington: Smithsonian Institution Press; 1993.
- Gouteux JP, Noireau F. The host preferences of Chrysops silacea and C. dimidiata. Diptera: Tabanidae in an endemic area of Loa loa in the Congo. Ann Trop Med Parasitol. 1989;83:167–72.PubMedGoogle Scholar
- Peters W, Garnham PCC, Rajapaksa N, Cheong WH, Cadigan FC. Malaria of the Orangutan in Borneo. Philos Trans R Soc Lond B Biol Sci. 1976;275:439–82. DOIPubMedGoogle Scholar
- Suleman MA, Johnson BJ, Tarara R, Sayer PD, Ochieng DM, Muli JM, An outbreak of poliomyelitis caused by poliovirus type I in captive black and white colobus monkeys (Colobus abyssinicus kikuyuensis) in Kenya. Trans R Soc Trop Med Hyg. 1984;78:665–9. DOIPubMedGoogle Scholar
- Goodall J. Population dynamics during a 15 year period in one community of free-living chimpanzees in the Gombe National Park, Tanzania. Z Tierpsychol. 1983;61:1–60.
- Mak JW, Cheong WH, Yen PK, Lim PK, Chan WC. Studies on the epidemiology of subperiodic Brugia malayi in Malaysia: problems in its control. Acta Trop. 1982;39:237–45.PubMedGoogle Scholar
- Rolland RM, Hausfater G, Marshall B, Levy SB. Antibiotic-resistant bacteria in wild primates: increased prevalence in baboons feeding on human refuse. Appl Environ Microbiol. 1985;49:791–4.PubMedGoogle Scholar
- Turner IM. Species loss in fragments of tropical rain forest: a review of the evidence. J Appl Ecol. 1996;33:200–9. DOIGoogle Scholar
- Wilson ME. Travel and the emergence of infectious diseases. Emerg Infect Dis. 1995;1:39–46. DOIPubMedGoogle Scholar
- Grenfell BT, Dobson AP, eds. Ecology of infectious diseases in natural populations. Cambridge: Cambridge University Press; 1995.
- Fleagle JG, Reed KE. Comparing primate communities: a multivariate approach. J Hum Evol. 1996;30:489–510. DOIGoogle Scholar
- Davies CR, Ayres JM, Dye C, Deane LM. Malaria infection rate of Amazonian primates increases with body weight and group size. Funct Ecol. 1991;5:655–62. DOIGoogle Scholar
- Freeland WJ. Pathogens and the evolution of primate sociality. Biotropica. 1976;8:12–24. DOIGoogle Scholar
- Loehle C. Social barriers to pathogen transmission in wild animal populations. Ecology. 1995;76:326–35. DOIGoogle Scholar
- Le Guenno B, Formentry P, Wyers M, Gounon P, Walker F, Boesch C. Isolation and partial characterization of a new strain of Ebola virus. Lancet. 1995;345:1271–444. DOIPubMedGoogle Scholar
- World Health Organization. Outbreak of Ebola haemorrhagic fever in Gabon officially declared over. Wkly Epidemiol Rec. 1996;71:125–6.
- Jezek Z, Arita I, Mutombo M, Dunn C, Nakano JH, Szczeniowski M. Four generations of probable person-to-person transmission of human monkeypox. Am J Epidemiol. 1986;123:1004–12.PubMedGoogle Scholar
- Fenner SS. Human monkeypox, a newly discovered human virus disease. In: Morse SS, editor. Emerging viruses. New York: Oxford University Press, Inc.; 1993. p. 176-183.
- Palmer AE. Herpesvirus simiae: historical perspective. J Med Primatol. 1987;16:99–130.PubMedGoogle Scholar
- Schrag SJ, Wiener P. Emerging infectious disease: what are the relative roles of ecology and evolution? Trends Ecol Evol. 1995;10:319–24. DOIGoogle Scholar
- Karesh WB, Cook RA. Applications of veterinary medicine to in situ conservation efforts. Oryx. 1995;29:244–52. DOIGoogle Scholar
- Hafner MS, Page RDM. Molecular phylogenies and host-parasite cospeciation: gophers and lice as a model system. Philos Trans R Soc Lond B Biol Sci. 1995;349:77–83. DOIPubMedGoogle Scholar
- Freeland WJ. Primate social groups as biological islands. Ecology. 1979;60:719–28. DOIGoogle Scholar
- Jin MJ, Rogers J, Phillips-Conroy JE, Allan JS, Desrosiers RC, Shaw GM, Infection of a yellow baboon with simian immunodeficiency virus from African green monkeys: evidence for cross-species transmission in the wild. J Virol. 1994;68:8454–60.PubMedGoogle Scholar
- Jolly CJ, Phillips-Conroy JE, Turner TR, Broussard S, Allan JS. SIV-agm incidence over two decades in a natural population of Ethiopian grivet monkeys (Cercopithecus aethiops aethiops). J Med Primatol. 1996;25:78–83.PubMedGoogle Scholar
- Rodriguez del Valle M, Quakyi IA, Amuesi J, Quaye JT, Nkrumah FK, Taylor DW. Detection of antigens and antibodies in the urine of humans with Plasmodium falciparum malaria. J Clin Microbiol. 1991;29:1236–42.PubMedGoogle Scholar
- Karesh WB, Smith F, Frazier-Taylor H. A remote method for obtaining skin biopsy samples. Conserv Biol. 1987;1:261–2. DOIGoogle Scholar
- Centers for Disease Control and Prevention. Tuberculosis in imported nonhuman primatesUnited States, June 1990-May 1993. MMWR Morb Mortal Wkly Rep. 1993;42:572–6.PubMedGoogle Scholar
- Tarara R, Suleman MA, Sapolsky R, Wabomba MJ, Else JG. Tuberculosis in wild olive baboons, Papio cynocephalus anubis (Lesson), in Kenya. J Wildl Dis. 1985;21:137–40.PubMedGoogle Scholar
- Clayton DH, Wolfe ND. The adaptive significance of self-medication. Trends Ecol Evol. 1995;8:60–3. DOIGoogle Scholar
- Robles M, Aregullin M, West J, Rodriguez E. Recent studies on the zoopharmacognosy, pharmacology and neurotoxicology of sesquiterpene lactones. Planta Med. 1995;61:199–203. DOIPubMedGoogle Scholar
- Hill AV, Yates SN, Allsopp CE, Gupta S, Gilbert SC, Lalvani A, Human leukocyte antigens and natural selection by malaria. Philos Trans R Soc Lond B Biol Sci. 1994;346:379–85. DOIPubMedGoogle Scholar
- Skolnick AA. Newfound genetic defect hints at clues for developing novel antimalarial agents. JAMA. 1993;269:1765. DOIPubMedGoogle Scholar
- Stephenson J. Findings on host resistance genes for infectious diseases are pointing the way to drugs, vaccines. JAMA. 1996;275:1464–5. DOIPubMedGoogle Scholar