Volume 10, Number 7—July 2004
Research
Recombinant Viruses and Early Global HIV-1 Epidemic
Figure 3
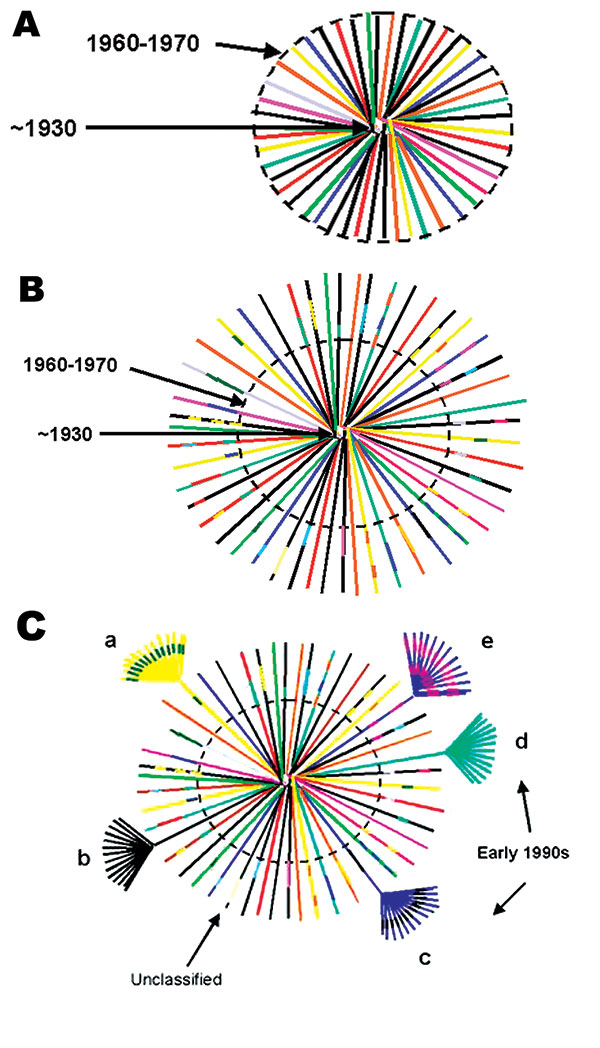
Figure 3. Hypothetical model of HIV-1, group M evolution. A. Star phylogeny representing the evolution of the ancestral HIV-1, group M virus that was able to adapt in humans and was transmitted among rural populations in Central Africa from approximately the 1930s (22). Over time, the viruses would have become increasingly genetically distinct from each other and the original parental strain. The dotted circle denotes the beginning of migration from these remote areas to cities in Central Africa (approximately 1960–1970). B. Recombinant lineages outside the dotted circle, represented by multicolored lines indicating mosaic viruses or genetic mixes of the circulating strains, would have been the result of population migration, urbanization, patterns of sexual activity, and medical practices (two of the oldest, fully characterized sequences, MAL [1985] and Z321 [1976], were both recombinant viruses from Zaire). C. Recombinant viruses would have continued to be generated and transmitted until introduced into high-risk populations, such as commercial sex workers, taxi drivers, commercial truck drivers, or long-distance truck drivers, and then rapidly transmitted within and between these social networks. Such high-risk social networks throughout central Africa were responsible for the rapid expansion of a relatively small number of evolving viruses, including recombinant strains, locally, regionally, and eventually globally. These epidemiologic groupings are represented as clusters of highly related strains at the end of a few HIV-1 lineages. Panel C shows what phylogenetic analysis of global HIV-1 strains collected in the early 1990s, when sequence characterization first began, and after being exported out of central Africa, would have looked like. The clusters of related sequences from founder viruses, which were disseminated globally, would have appeared as subtypes or clades, arbitrarily labeled a-e. Occasionally strains that were not widely expanded were identified and designated as unclassifiable. From this hypothetical modeling, and the high numbers of recombinant strains, it seems unlikely that only pure subtypes were exported from this region of Africa to establish mini-epidemics in other countries. Therefore, at least some of what we currently define as pure subtypes most likely arose from recombinant genomes originally generated somewhere in central Africa.
References
- Vidal N, Peeters M, Mulanga-Kabeya C, Nzilambi N, Robertson D, Ilunga W, Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J Virol. 2000;74:10498–507. DOIPubMedGoogle Scholar
- Murphy E, Korber B, Georges-Courbot MC, You B, Pinter A, Cook D, Diversity of V3 region sequences of human immunodeficiency viruses type 1 from the central African Republic. AIDS Res Hum Retroviruses. 1993;9:997–1006. DOIPubMedGoogle Scholar
- Yang C, Dash B, Hanna SL, Frances HS, Nzilambi N, Colebunders RC, Predominance of HIV type 1 subtype G among commercial sex workers from Kinshasa, Democratic Republic of Congo. AIDS Res Hum Retroviruses. 2001;17:361–5. DOIPubMedGoogle Scholar
- de Leys R, Vanderborght B, vanden Haesevelde M, Heyndrickx L, van Geel A, Wauters C, et al. Isolation and partial characterization of an unusual human immunodeficiency retrovirus from two persons of west-central Africa origin. J Virol. 1990;64:1207–16.PubMedGoogle Scholar
- Gurtler LG, Hauser PH, Eberle J, von Brunn A, Knapp S, Zekeng L, A new subtype of human immunodeficiency virus type 1 (MVP-5180) from Cameroon. J Virol. 1994;68:1581–5.PubMedGoogle Scholar
- Simon F, Mauclere P, Roques P, Loussert-Ajaka I, Muller-Trutwin MC, Saragosti S, Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat Med. 1998;4:1032–7. DOIPubMedGoogle Scholar
- Charneau P, Borman AM, Quillent C, Guetard D, Chamaret S, Cohen J, Isolation and envelope sequence of a highly divergent HIV-1 isolate: definition of a new HIV-1 group. Virology. 1994;205:247–53. DOIPubMedGoogle Scholar
- Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Origin of HIV-1 in chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–41. DOIPubMedGoogle Scholar
- Hahn GH, Shaw GM, De Cock KM, Sharp PM. AIDS as a zoonosis: scientific and public health implications. Science. 2000;287:607–14. DOIPubMedGoogle Scholar
- Mann JM, Francis H, Quinn T, Asila PK, Bosenge N, Nzilambi N, Surveillance for AIDS in a central African city, Kinshasa, Zaire. JAMA. 1986;255:3255–9. DOIPubMedGoogle Scholar
- Nzilambi N, De Cock KM, Forthal DN, Francis H, Ryder RW, Malbe I, The prevalence of infection with human immunodeficiency virus over a 10-year period in rural Zaire. N Engl J Med. 1988;318:276–9. DOIPubMedGoogle Scholar
- Mulanga-Kabeya C, Nzilambi N, Edidi B, Minlangu M, Tshimpaka T, Kambembo L, Evidence of stable HIV seroprevalences in selected populations in the Democratic Republic of the Congo. AIDS. 1998;12:905–10. DOIPubMedGoogle Scholar
- Mann JM, Nzilambi N, Piot P, Bosenge N, Kalala M, Francis H, HIV infection and associated risk factors in female prostitutes in Kinshasa, Zaire. AIDS. 1988;2:249–54. DOIPubMedGoogle Scholar
- N’Galy B, Ryder RW, Bila K, Mwandagalirwa G, Colebunders RL, Francis H, Human immunodeficiency virus infection among employees in an African hospital. N Engl J Med. 1988;319:1123–7. DOIPubMedGoogle Scholar
- Ellenberger DL, Pieniazek D, Nkengasong J, Luo C-C, Maurice C, Janini M, Genetic analysis of human immunodeficiency virus in Abidjan, Cote d’Ivoire reveals predominance of HIV-1 subtype A and introduction of subtype G. AIDS Res Hum Retroviruses. 1999;15:3–9. DOIPubMedGoogle Scholar
- Schochetman GS, Subbarao S, Kalish ML. Methods for studying genetic variation of the human immunodeficiency virus (HIV). In: Adolph KA, editor. Viral genome methods. Boca Raton (FL): CRC Press; 1996. p. 25–41.
- Yang C, Pieniazek D, Owen SM, Fridlund C, Nkengasong J, Mastro TD, Detection of phylogenetically diverse human immunodeficiency virus type 1 groups M and O from plasma by using highly sensitive and specific generic primers. J Clin Microbiol. 1999;37:2581–6.PubMedGoogle Scholar
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–8. DOIPubMedGoogle Scholar
- Swofford DL. PAUP* Phylogenetic Analysis Using Parsimony (*and Other Methods) Version 4. Sunderland (MA): Sinauer Associates; 2003.
- Iglesias-Sanchez MJ, Lopez-Galindez C. Analysis, quantification, and evolutionary consequences of HIV-1 in vitro recombination. Virology. 2002;304:392–402. DOIPubMedGoogle Scholar
- Anderson JP, Rodrigo AG, Learn GH, Madan A, Delahunty C, Coon M, Testing the hypothesis of a recombinant origin of human immunodeficiency virus type 1 subtype E. J Virol. 2000;74:10752–65. DOIPubMedGoogle Scholar
- Oucho JO, Gould WTS. Chapter 7: internal migration, urbanization, and population distribution. In: Foote KA, Hill KH, Martin LG, editors. Demographic change in sub-Saharan Africa. Washington (DC): National Academy Press; 1993. p. 256–96.
- Mann JM, Francis H, Davachi F, Baudoux P, Quinn TC, Nzilambi N, Risk factors for human immunodeficiency virus seropositivity among children 1 to 24 months old in Kinshasa, Zaire. Lancet. 1986;2:654–7. DOIPubMedGoogle Scholar
- Mann JM, Francis H, Davachi F, Baudoux P, Quinn TC, Nzilambi N, Human immunodeficiency virus seroprevalence in pediatric patients 2 to 14 years of age at Mama Yemo Hospital, Kinshasa, Zaire. Pediatrics. 1986;78:673–7.PubMedGoogle Scholar
- Mann JM, Francis H, Quinn TC, Bila K, Asila PK, Bosenge N, HIV seroprevalence among hospital workers in Kinshasa, Zaire. JAMA. 1986;256:3099–102. DOIPubMedGoogle Scholar
- Greenberg AE, Nguyen-Dinh P, Mann JM, Kobote N, Colebunders RL, Francis H, The association between malaria, blood transfusions, and HIV seropositivity in a pediatric population in Kinshasa, Zaire. JAMA. 1988;259:545–9. DOIPubMedGoogle Scholar
- Jager H, N’Galy B, Perriens J, Nseka K, Davachi F, Kabeya CM, Prevention of transfusion-associated HIV transmission in Kinshasa, Zaire: HIV screening is not enough. AIDS. 1990;4:571–4. DOIPubMedGoogle Scholar
- N’tita I, Mulanga K, Dulat C, Lusamba D, Rehle T, Korte R, Risk of transfusion-associated HIV transmission in Kinshasa, Zaire. AIDS. 1991;5:437–9. DOIPubMedGoogle Scholar
- Gaschen B, Taylor J, Yusim K, Foley B, Gao F, Lang D, Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–60. DOIPubMedGoogle Scholar
- Nickle DC, Jensen MA, Gottlieb GS, Shriner D, Learn GH, Rodrigo AG, Consensus and ancestral state HIV vaccines. Science. 2003;299:1515–8. DOIPubMedGoogle Scholar
- Ramos A, Hu DJ, Nguyen L, Phan KO, Vanichseni S, Promadej N, Intersubtype human immunodeficiency virus type 1 superinfection following seroconversion to primary infection in two injection drug users. J Virol. 2002;76:7444–52. DOIPubMedGoogle Scholar
- Schierup MH, Hein J. Consequences of recombination on traditional phylogenetic analysis. Genetics. 2000;156:879–91.PubMedGoogle Scholar
- Wain-Hobson S, Renoux-Elbe C, Vartanian J-P, Meyerhans A. Network analysis of human and simian immunodeficiency virus sequence sets reveals massive recombination resulting in shorter pathways. J Gen Virol. 2003;84:885–95. DOIPubMedGoogle Scholar
- Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–96. DOIPubMedGoogle Scholar