Volume 16, Number 12—December 2010
CME ACTIVITY - Dispatch
Oseltamivir-Resistant Pandemic (H1N1) 2009 Virus, South Korea
Introduction

Medscape, LLC is pleased to provide online continuing medical education (CME) for this journal article, allowing clinicians the opportunity to earn CME credit. This activity has been planned and implemented in accordance with the Essential Areas and policies of the Accreditation Council for Continuing Medical Education through the joint sponsorship of Medscape, LLC and Emerging Infectious Diseases. Medscape, LLC is accredited by the ACCME to provide continuing medical education for physicians. Medscape, LLC designates this educational activity for a maximum of 0.25 AMA PRA Category 1 Credits™. Physicians should only claim credit commensurate with the extent of their participation in the activity. All other clinicians completing this activity will be issued a certificate of participation. To participate in this journal CME activity: (1) review the learning objectives and author disclosures; (2) study the education content; (3) take the post-test and/or complete the evaluation at www.medscapecme.com/journal/eid; (4) view/print certificate.
Learning Objectives
Upon completion of this activity, participants will be able to:
- Describe the pattern and characteristics of resistance seen with antiviral agents in pandemic (H1N1) 2009 in South Korea.
Medscape CME Editor
Thomas J. Gryczan, MS, Technical Writer/Editor, Emerging Infectious Diseases. Disclosure: Thomas J. Gryczan, MS, has disclosed no relevant financial relationships.
Medscape CME Author
Desiree Lie, MD, MSEd, Clinical Professor; Director of Research and Faculty Development, Department of Family Medicine, University of California, Irvine at Orange. Disclosure: Désirée Lie, MD, MSEd, has disclosed the following relevant financial relationship: served as a nonproduct speaker for "Topics in Health" for Merck Speaker Services.
Authors
Disclosures: Hwajung Yi, PhD; Joo-Yeon Lee, PhD; Eun-Hye Hong, BS; Mi-Seon Kim, BS; Donghyok Kwon, MS; Jang-Hoon Choi, MS; Woo-Young Choi, PhD; Ki-Soon Kim, PhD; Jong-Koo Lee, MD; Hee-Bok Oh, PhD; and Chun Kang, MS, have disclosed no relevant financial relationships.
Abstract
To identify oseltamivir resistance, we analyzed neuraminidase H275Y mutations in samples from 10 patients infected with pandemic (H1N1) 2009 virus in South Korea who had influenza that was refractory to antiviral treatment with this drug. A neuraminidase I117M mutation that might influence oseltamivir susceptibility was detected in sequential specimens from 1 patient.
Since April 2009, pandemic (H1N1) 2009 has spread worldwide and caused the first influenza pandemic of the 21st century. Pandemic (H1N1) 2009 virus initially showed resistance to amantadine but susceptibility to oseltamivir (1). Thereafter, 285 cases of oseltamivir-resistant pandemic viral infection were reported worldwide on April 14, 2010 (2). However, information is limited about oseltamivir-resistant pandemic influenza in South Korea. Monitoring of community circulation of oseltamivir-resistant viruses has not yet detected any evidence of oseltamivir resistance in South Korea. To identify these viruses, we conducted specific surveillance of antiviral drug–resistant infection in patients whose illness did not resolve after antiviral treatment.
The study was reviewed and approved by ethics committees of relevant institutions and hospitals. After patients provided informed consent, we obtained >150 clinical specimens from patients in various hospitals in South Korea. Respiratory specimens (>60% nasopharyngeal swab specimens) were obtained during October 2009–January 2010 from patients whose illness had been clinically refractory to antiviral treatment since October 2009.
Viral RNAs were isolated from specimens of 10 patients (Table 1) by using the QIAamp viral RNA Mini Kit (QIAGEN, Crawley, UK). PCR products of the neuraminidase (NA) and matrix 2 (M2) genes were generated by reverse transcription–PCR with primers for NA (forward: 5′-AAATTAACGGGCAATTCCTCTCT-3′; reverse: 5′-CCGAAAATCCCACTGCATATGTAT-3′) and M2 (forward: 5′-CTAGCTCCAGTGCTGGTCTGA-3′; reverse: 5′-CTCAGGCACTCCTTCCGTAGA-3′). DNA sequences of NA and M2 reverse transcription–PCR products were analyzed by using the Big-Dye Terminator Sequencing Reaction Kit and an ABI 3700 DNA analyzer (Applied Biosystems, Foster City, CA, USA). A total of 58 NA sequences and 52 M2 sequences were obtained and analyzed by using the sequence analysis tool in the Influenza Sequences and Epitopes Database for detecting oseltamivir-resistance mutations (3).
Ten patients were detected who had oseltamivir-resistant pandemic (H1N1) 2009 virus with the H275Y substitution in viral NA (Table 2). Oseltamivir resistance was associated with oseltamivir treatment on the basis of H275Y changes from the oseltamivir-sensitive genotypes to oseltamivir-resistant genotypes of viral NA in consecutive samples from the same patient. Furthermore, a novel NA (I117M) substitution that may be associated with oseltamivir resistance (4,5) was detected in specimens from 1 patient (patient G) who had myelodysplasia and received oseltamivir and peramivir (Table 1, Table 2).
In addition, we cultured viral isolates from clinical specimens (patients A and C) and evaluated antiviral susceptibility by measuring the dose of oseltamivir and zanamivir required for 50% inhibition (IC50) of NA activity. One isolate of pandemic (H1N1) 2009 virus with an oseltamivir-sensitive genotype (H275 in its NA) was susceptible to oseltamivir (IC50 1.18 nmol/L) and zanamivir (IC50 0.42 nmol/L). Viral isolates from patients A and C with an oseltamivir-resistant genotype (Y275 in NA) were resistant only to oseltamivir (IC50 713.2 nmol/L and 359.4 nmol/L, respectively). Susceptibility to zanamivir was not altered whether NA contained Y275 or H275 (IC50 0.13 nmol/L and 0.78 nmol/L, respectively).
Patients with oseltamivir-resistant pandemic (H1N1) 2009 were treated during hospitalization with oseltamivir alone or with a combination of other antiviral drugs (Table 2). Active surveillance that evaluated spread of oseltamivir-resistant viral infections among hospital staff, family members, and other patients who had contacted with or cared for the patients showed no evidence of virus transmission in the hospitals.
During pandemic (H1N1) 2009, oseltamivir-resistant viral infections were found mainly in immunocompromised patients who were treated with antiviral drugs and chemoprophylaxis (6). In our study, genetic characteristics of oseltamivir resistance in virus isolates from all 10 patients showed the H275Y substitution in NA, resulting in phenotypic resistance to oseltamivir but not to zanamivir. In addition, a novel NA mutation (I117M) was detected in virus from patient G (Table 1, Table 2). The NA 117 residue is near R118 and may have an effect on this residue, 1 of 3 arginine residues that bind the carboxylate region of the sialic acid substrate (5,7). This finding implies that changes at NA residue 117 may influence oseltamivir susceptibility of the virus. For example, the NA I117V mutation in avian influenza virus (H5N1) was reported to increase resistance to oseltamivir (5,6). The 117 residue is also highly conserved in all N1 typed viruses (5). These results indicate that the I117M substitution in NA of pandemic (H1N1) 2009 virus may influence oseltamivir resistance of this virus.
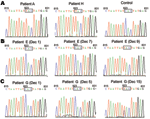
Multiple specimens were available from patients D, E, and G for analyzing antiviral resistance mutations in NA. Analysis of viruses from these patients showed that the H275Y mutation emerged in the later stages of viral infection, during oseltamivir treatment; infective viruses initially had an oseltamivir-sensitive genotype (H275) (Table 2; Figure).
A mixture of oseltamivir-sensitive (H275 in NA) and oseltamivir-resistant (Y275 in NA) viruses was observed in the second specimen from patient E and the third specimen from patient G (Figure). These specimens showed double signals (nucleotides T and C) for TAC and CAC encoding tyrosine (Y) and histidine (H), respectively. There are 2 possible explanations for mixtures of oseltamivir-resistant and oseltamivir-sensitive viral genotypes. The first explanation is reinfection with Y275 virus after a previous infection with H275 virus. The second explanation is an H275Y mutation caused by selective pressure from oseltamivir treatment. Our molecular epidemiologic data (Table 2) support the second explanation that selective pressure from oseltamivir treatment caused the H275Y substitution in NA because sequential viruses from the same patient had identical HA sequences in the absence of additional infection (8).
Multiple specimens from the same patient showed identical hemagglutinin sequences despite amino acid changes (H to Y) at position 275 of NA (hemagglutinin sequences of patients D, E, and G) (Table 2). This finding indicates that temporally sequential viruses from a patient were likely generated from the same viral progenitor without further viral infection. Conversely, our findings also support absence of epidemiologic links among current cases of oseltamivir resistance, given that viruses from 9 patients in this study had their own signature hemagglutinin amino acid sequences (Table 2).
Spreading and clustering of oseltamivir-resistant strains in the general population has not been observed in South Korea. However, in the present study, a comparatively high rate of emergence (19%) of oseltamivir-resistant viral infections was observed in unhealthy patients treated with antiviral drugs. We also detected a novel substitution (I117M) in NA, which might influence susceptibility to NA inhibitors. Our findings demonstrate that oseltamivir-resistant quasispecies of pandemic (H1N1) 2009 viruses can be generated. This suggestion indicates that continuous surveillance is required to evaluate emergence and circulation of drug-resistant pandemic (H1N1) 2009 viruses and possible reassortment with other viruses that have oseltamivir resistance.
Dr Yi is a research scientist in the Division of Influenza Viruses, Center for Infectious Diseases, National Institute of Health, Centers for Disease Control and Prevention, Seoul, South Korea. His research interests are vaccine development, antiviral resistance, and immunologic properties of influenza viruses.
Acknowledgment
This study was supported by an intramural research fund (code# 2010-N43003-00) of the Korea Centers for Disease Control and Prevention.
References
- Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team, Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, et al. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med. 2009;360:2605–15. DOIPubMedGoogle Scholar
- World Health Organization. Weekly update on oseltamivir resistance to pandemic influenza A (H1N1) 2009 update 99, April [cited 2010 Aug 31]. http://www.who.int/csr/disease/swineflu/oseltamivirresistant2010416.pdf
- Yang IS, Lee JY, Lee JS, Mitchell WP, Oh HB, Kang C, Influenza sequence and epitope database. Nucleic Acids Res. 2009;37:D423–30. DOIPubMedGoogle Scholar
- Govorkova EA, Ilyushina NA, McClaren JL, Naipospos TS, Douangngeun B, Webster RG. Susceptibility of highly pathogenic H5N1 influenza viruses to the neuraminidase inhibitor oseltamivir differs in vitro and in a mouse model. Antimicrob Agents Chemother. 2009;53:3088–96. DOIPubMedGoogle Scholar
- Hurt AC, Selleck P, Komadina N, Shaw R, Brown L, Barr IG. Susceptibility of highly pathogenic A (H5N1) avian influenza viruses to the neuraminidase inhibitors and adamantanes. Antiviral Res. 2007;73:228–31. DOIPubMedGoogle Scholar
- Update on oseltamivir resistant pandemic A (H1N1) 2009 influenza virus: January 2010. Wkly Epidemiol Rec. 2009;85:37–40.PubMedGoogle Scholar
- Russell RJ, Haire LF, Stevens DJ, Collins PJ, Lin YP, Blackburn GM, The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature. 2006;443:45–9. DOIPubMedGoogle Scholar
- Gubareva LV, Novikov DV, Hayden FG. Assessment of hemagglutinin sequence heterogeneity during influenza virus transmission in families. J Infect Dis. 2002;186:1575–81. DOIPubMedGoogle Scholar
Figure
Tables
Follow Up
To obtain credit, you should first read the journal article. After reading the article, you should be able to answer the following, related, multiple-choice questions. To complete the questions and earn continuing medical education (CME) credit, please go to www.medscapecme.com/journal/eid. Credit cannot be obtained for tests completed on paper, although you may use the worksheet below to keep a record of your answers. You must be a registered user on Medscape.com. If you are not registered on Medscape.com, please click on the New Users: Free Registration link on the left hand side of the website to register. Only one answer is correct for each question. Once you successfully answer all post-test questions you will be able to view and/or print your certificate. For questions regarding the content of this activity, contact the accredited provider, CME@medscape.net. For technical assistance, contact CME@webmd.net. American Medical Association's Physician's Recognition Award (AMA PRA) credits are accepted in the US as evidence of participation in CME activities. For further information on this award, please refer to http://www.ama-assn.org/ama/pub/category/2922.html. The AMA has determined that physicians not licensed in the US who participate in this CME activity are eligible for AMA PRA Category 1 Credits™. Through agreements that the AMA has made with agencies in some countries, AMA PRA credit is acceptable as evidence of participation in CME activities. If you are not licensed in the US and want to obtain an AMA PRA CME credit, please complete the questions online, print the certificate and present it to your national medical association.
Oseltamivir-Resistant Pandemic (H1N1) 2009 Virus, South Korea
Medscape CME Questions
1. Which of the following best describes the pattern of oseltamivir resistance seen with pandemic (H1N1) 2009 virus in South Korea?
A. Resistance was seen synchronous with amantadine resistance
B. Resistance was seen in the community
C. Resistance was seen in hospitalized patients
D. Resistance was seen only in immunosuppressed patients
2. A 45-year-old man from South Korea with pandemic (H1N1) 2009 infection is found to have resistance to oseltamivir. Which of the following is the most likely outcome of this resistance?
A. Increased transmission of virus to staff and family
B. Higher risk for resistance in the community
C. Increased length of hospital stay
D. None of the above
Activity Evaluation
| 1. The activity supported the learning objectives. | ||||
| Strongly Disagree |
Strongly Agree
|
|||
|
1
|
2
|
3
|
4
|
5
|
| 2. The material was organized clearly for learning to occur. | ||||
| Strongly Disagree |
Strongly Agree
|
|||
|
1
|
2
|
3
|
4
|
5
|
| 3. The content learned from this activity will impact my practice. | ||||
| Strongly Disagree |
Strongly Agree
|
|||
|
1
|
2
|
3
|
4
|
5
|
| 4. The activity was presented objectively and free of commercial bias. | ||||
| Strongly Disagree |
Strongly Agree
|
|||
|
1
|
2
|
3
|
4
|
5
|
Related Links
Table of Contents – Volume 16, Number 12—December 2010
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Chun Kang, Division of Influenza Viruses, Center for Infectious Diseases, National Institute of Health, Korea Centers for Disease Control and Prevention, Nokbeon-dong, Eunpyeong-gu, Seoul, 122–701, South Korea
Top