Volume 16, Number 4—April 2010
Research
Livestock-associated Methicillin-Resistant Staphylococcus aureus Sequence Type 398 in Humans, Canada
Abstract
Rates of colonization with livestock-associated methicillin-resistant Staphylococcus aureus (MRSA) sequence type 398 have been high for pigs and pig farmers in Canada, but prevalence rates for the general human population are unknown. In this study, 5 LA-MRSA isolates, 4 of which were obtained from skin and soft tissue infections, were identified from 3,687 tested MRSA isolates from persons in Manitoba and Saskatchewan, Canada. Further molecular characterization determined that these isolates all contained staphylococcal cassette chromosome (SCC) mecV, were negative for Panton-Valentine leukocidin, and were closely related by macrorestriction analysis with the restriction enzyme Cfr91. The complete DNA sequence of the SCCmec region from the isolate showed a novel subtype of SCCmecV harboring clustered regularly interspaced short palindromic repeats and associated genes. Although prevalence of livestock-associated MRSA seems to be low for the general population in Canada, recent emergence of infections resulting from this strain is of public health concern.
High prevalence of colonization with livestock-associated (LA) methicillin-resistant Staphylococcus aureus (MRSA) sequence type (ST) 398 among pigs and pig farmers was first reported in the Netherlands (1) and has since been identified in Canada (2) and the United States (3). In Canada, this LA-MRSA strain was identified in pigs and pig farmers in southwestern Ontario, where prevalence of MRSA colonization was 24.9% (71/285) and 20% (5/25), respectively (2). In the United States, nasal samples from 20 production system workers and 299 swine from 2 farms in Illinois and Iowa showed that 45% (9/20) and 49% (147/299), respectively, were colonized with LA-MRSA (3). Despite such high prevalence of MRSA colonization on these tested farms, to our knowledge, no human or animal infections resulting from LA-MRSA strains have been reported in North America.
To determine whether LA-MRSA has recently emerged in the general population of Canada, we identified human infections and colonizations associated with the LA-MRSA strain in Canada and molecularly characterized the isolates. We also identified a novel staphylococcal cassette chromosome (SCC) mecV subtype harboring clustered regularly interspaced short palindromic repeats (CRISPR) and CRISPR-associated genes (cas).
A convenience sample, totaling 2,358 MRSA isolates from human specimens, was submitted to the National Microbiology Laboratory (NML) for spa typing, as described (4,5). During January 2007–October 2008, the Saskatchewan Disease Control Laboratory submitted 2,008 specimens; during October 2007–August 2008, the Cadham Provincial Laboratory in Manitoba submitted 350 specimens. An additional 1,329 isolates from human specimens were spa typed by the Saskatchewan Disease Control Laboratory.
Given the client base of the Cadham Provincial Laboratory, most of these isolates would have originated from colonized and infected persons living in the community or in personal-care homes or from persons hospitalized in smaller community hospitals, whereas, for surveillance purposes, the Saskatchewan Disease Control Laboratory receives isolates from all colonized and infected persons across the province. Detailed information regarding why cultures were taken (e.g., screening admissions, outbreak investigations) and other clinical and epidemiologic data were limited. Isolates typed in this study represented ≈17% of all MRSA isolates from persons in Manitoba and ≈66% of all MRSA isolates from persons in Saskatchewan within the study period. An additional isolate was sent to the NML from Sunnybrook Health Sciences Centre in Ontario for reference purposes.
Isolates with spa types associated with ST398 were confirmed by multilocus sequence typing; tested for Panton-Valentine leukocidin toxin, mecA, and nuc genes; and typed for SCCmec as described (6–9). Pulsed-field gel electrophoresis (PFGE) of SmaI- or Cfr91-digested genomic DNA was conducted as described (10). Antimicrobial drug susceptibility testing was conducted by using standard broth microdilution panels according to Clinical and Laboratory Standards Institute guidelines (11). Breakpoints for fusidic acid and mupirocin resistance, which were not provided in the guidelines, were as described (12,13).
A fosmid library was constructed by cloning sheared genomic DNA from S. aureus isolate 08 BA 02176 into the pCC2FOS vector. The fosmid clones were screened by PCR to identify specific genes orfX, mecA, and the chromosomal region located downstream of SCCmec. Fosmid clone 1G1 was identified and contained the entire SCCmec region of the 08 BA 02176 isolate. Colonies were prepared by using the CopyControl Fosmid Library Production Kit (Epicenter Biotechnologies, Madison, WI, USA) according to the manufacturer’s instructions. Fosmid DNA was column purified by using a QIAGEN Plasmid Mini Kit (QIAGEN, Valencia, CA, USA).
DNA sequencing was performed on the ABI3730xl genetic analyzer (Applied Biosystems, Foster City, CA, USA). Staden (Pregap4) software was used to prepare trace data for sequence assembly (14). Sequencing reads were assembled by using the Staden Gap4 program. Gap closure was achieved by primer walking and long-range PCR. Specific primers were designed near the ends of neighboring contigs (contiguous sequence of DNA created by overlapping sequenced fragments of a chromosome), and PCRs were performed with chromosomal template DNA. Regions containing putative frameshifts and point mutations were resequenced to verify the fidelity of the sequence.
Annotation and data mining of the S. aureus 08 BA 02176 1G1 fosmid clone sequence were performed by using the GenDB version 2.2 annotation tool (15). Putative protein coding sequences were determined according to coding sequence predictions of Glimmer, which is integrated into the GenDB package. Similarity searches were performed by using BLASTN and BLAST2P (www.ncbi.nlm.nih.gov/blast/Blast.cgi) against the nonredundant nucleotide and protein databases, respectively. Additionally, a BLAST2P search was performed against the databases nr (ftp://ftp.ncbi.nlm.nih.gov/blast/db/), SWISS-PROT (www.expasy.ch/sprot/), and KEGG-Genes (ftp://ftp.genome.jp/pub/kegg/genes/); the protein family databases Pfam (http://pfam.sanger.ac.uk/) and TIGRFAM (www.jcvi.org/cms/research/projects/tigrfams/overview/); and predictive signal peptide (Signal P [www.cbs.dtu.dk/services/SignalP/]) and transmembrane helix analysis (TMHMM [www.cbs.dtu.dk/services/TMHMM/]), the nonredundant database on protein level. An automatic functional annotation was followed by a manual annotation of each predicted gene.
LA-MRSA Characterization
A total of 3,687 MRSA isolates were examined; 5 contained ST398-associated spa types (4 t034 and 1 t1250). The additional isolate submitted to NML by Sunnybrook Health Sciences Centre in Ontario, isolate T40929, also contained a t034 spa type. Further molecular characterization of these 6 isolates determined that they were all ST398, SCCmecV, and negative for the Panton-Valentine leukocidin–encoding genes (Table 1). Of the 6 isolates, 5 were resistant to tetracycline, but all were susceptible to the other 12 antimicrobial drugs tested (Table 2).
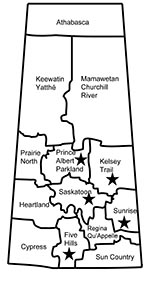
From the surveillance in Manitoba and Saskatchewan, patient information was limited and showed no geographic links (all 5 persons resided in different health regions but were all within the southeastern portion of Saskatchewan) (Figure 1). Of the 5 isolates, 4 were obtained from infected persons (average age 67.8 years, range 51–79 years) (Table 1). The earliest identified LA-MRSA isolate (08 BA 2176) associated with an infection was obtained from a postoperative surgical site. Further follow-up was not possible because of the patient’s health problems. This patient is unlikely to have had any recent direct contact with livestock because she had been confined to her home with limited mobility for several years before her hospitalization. Additional nasal swabs from this patient remained positive for this strain for at least 7 months. Additional clinical and epidemiologic information for the remaining 3 patients with skin and soft tissue or wound infections were limited (Table 1).
The isolate submitted to the NML by Sunnybrook Health Sciences Centre, outside the surveillance program, was from a 59-year-old man from Ontario. He had been hospitalized in December 2007 for treatment of metastatic squamous cell carcinoma of the larynx. In the previous year, he had undergone a total laryngectomy, neck node dissection, and tracheostomy. A MRSA isolate was recovered from screening specimens from his nose and the tracheostomy site that had no indication of infection. He was unaware of any animal contact and had no history of exposure to pigs or pig farms. A review of the medical records and standard epidemiologic investigations determined that this was not a nosocomial or healthcare-associated isolate.
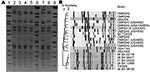
The 6 LA-MRSA isolates were nontypeable by PFGE using SmaI. However, PFGE using the neoschizomer Cfr91 showed that the 6 LA-MRSA isolates were closely related (Figure 2, panel A). Control MRSA strains digested individually with either Cfr91 or SmaI showed no differences in fingerprint banding patterns when the 2 enzymes were compared (data not shown), which enabled comparisons of the PFGE patterns obtained for the LA-MRSA isolates with those of other epidemic MRSA strains from hospitals and communities in Canada. No close relatedness was found (>7 bands difference; <80% similarity) between the LA-MRSA isolates and any other epidemic MRSA strain circulating in Canada (Figure 2, panel B).
SCCmec Characterization
DNA sequencing of the entire SCCmec element from isolate 08 BA 02176 showed a 32,369-bp element integrated at the 3′ end of orfX containing 30 putative open reading frames (ORFs) (Figure 3; Table 3). This element carried a class C2 mec complex, which putatively contained a nonfunctional IS431 transposase and a type 5 ccr gene complex (ccrC2). Other than mecA, no additional antimicrobial drug resistance genes were identified within this element.
The first unique feature of this SCCmecV element included 3 ORFs in the J3 region sharing high sequence identity with ORFs from S. epidermidis RP62A (GenBank accession no. CP000029), which included an ADP-ribosylglycohydrolase, a permease for cytosine/purines, and a ribokinase (Table 3). A second unique feature was a CRISPR array, identified by using CRISPRFinder (16), in the J1 region, which appears to have replaced the type 1 restriction modification system (hsdR, hsdS, hsdM) through recombination. The CRISPR array (1,107 bp) contained a 36-bp direct repeat consensus and 15 spacers of variable sequence and length (33–38 bp) (Figure 3). Downstream of this CRISPR array was a combination of putative CRISPR-associated (cas) genes, sharing sequence identity with those previously identified in S. epidermidis RP62A. This array was followed by a second questionable CRISPR array (183 bp) containing a 38-bp direct repeat consensus and 2 spacers of variable sequence (Figure 3; Table 3).
Design of primers spanning the entire SCCmec element was based on the DNA sequence obtained from 08 BA 02176 (Figure 3; Table 4). PCR of these select regions produced amplicons of expected size for 3 additional LA-MRSA isolates (07 BA 06477, 08 BA 13895, 08 BA 22334) but were negative for some of the J1 and J3 regions in 08 BA 08100 and T40929 (Table 4).
The high prevalence of LA-MRSA colonization of pigs and pig farmers in Canada (2) and the United States (3) and this report of human infections suggest that this LA-MRSA strain from Canada poses potential public and occupational health concern in North America. This strain has been associated with various types of infections in pigs (17,18) and humans (19,20) and is transmissible from animal patients to veterinary workers (21), healthcare workers (22), and family members (1). Evidence also suggests that this strain might be spreading from animals to the environment, which may facilitate the colonization or infection of persons who are not involved in animal husbandry (23). Whereas in 2006 in the Netherlands LA-MRSA accounted for >20% of all MRSA isolated (24), carriage of this strain in the general population of 2 provinces in Canada (Manitoba and Saskatchewan) appears rare (0.14%). This difference could be attributed to the substantially higher density of pigs in the Netherlands (1,244 pigs/km2) than in Manitoba (55 pigs/km2), Saskatchewan (6 pigs/km2), and Ontario (91 pigs/km2) (www.agriculture.gov.sk.ca/Pig_Densities). It is also plausible that the much lower proportions of LA-MRSA in Canada, relative to a country with low MRSA endemicity such as the Netherlands, is attributable to competition with other highly successful human epidemic MRSA clones circulating in Canada, including CMRSA2 (USA200/800), CMRSA7 (USA400), and CMRSA10 (USA300) (25,26).
The tested LA-MRSA isolates were highly susceptible to most classes of antimicrobial drugs, except β-lactams and tetracyclines, the latter of which has been attributed to its high usage in animal husbandry (27). The complete sequence of the SCCmec region showed a novel SCCmecV subtype sharing sequence identity in its J1 and J3 regions with chromosomal genes in the S. epidermidis RP62A chromosome (GenBank accession no. CP000029), including a CRISPR system. CRISPRs and associated cas genes are present in many other bacterial (≈40%) and archaeal (≈90%) genomes (28,29) and have been shown to be involved in sequence-directed immunity against phages (30,31) and plasmids (32). The resistance to plasmids and phages encoded by this system could explain why many of these ST398-MRSA-V strains contain fewer antimicrobial drug resistance genes and phage-encoded virulence factors than do other epidemic MRSA strains (33,34). The origin of this CRISPR system is unknown, but the propagation of CRISPR loci throughout prokaryote genomes has been proposed to occur through horizontal gene transfer by conjugation of megaplasmids >40 kb (35). Because the CRISPR system identified in this study is encoded within a putative mobile genetic element, we propose that an additional mechanism of mobilization to other methicillin-susceptible Staphylococcus spp. is plausible.
This novel subtype of SCCmecV was found in only 4 of the 6 LA-MRSA isolates identified in this study. One isolate not containing this novel SCCmec subtype (08 BA 08100) could also be distinguished by a different but closely related spa type (t1250) (Table 1) and variant PFGE fingerprint (Figure 2) when compared with the other LA-MRSA isolates, which suggests that at least 2 epidemiologically different strains of LA-MRSA circulate in Saskatchewan. The other LA-MRSA isolate that did not contain this novel SCCmec element was obtained in Ontario. However, this isolate was the same spa type (t034) and was closely related, according to PFGE, to the LA-MRSA isolates identified in Saskatchewan. Therefore, in addition to PFGE and spa typing, SCCmec subtyping could provide a useful epidemiologic tool for surveillance, outbreak investigations, or traceability studies of this emerging strain. For detection of this SCCmecV subtype (tentatively designated V.2.1.2; Vb), we propose using primer set 1 (spanning orfX into Sk02 in the J3 region) and primer set 7 (spanning Sk20 into cas1 in the J1 region) (Table 4).
Visual comparison of PFGE fingerprints from this study with those reported from patients from the Dominican Republic and the United States (northern Manhattan, New York, NY) (36), showed substantial variations in fingerprint patterns, as well as related but different spa types. These variations suggest further molecular and geographic diversity of these LA-MRSA strains on a global scale.
Because cases of LA-MRSA infections have only recently been identified in Canada, additional surveillance efforts are required to monitor the emergence and clinical relevance of this MRSA strain in Canada, including communities, the environment, livestock, farmers, and production facility workers. Whether these strains pose a major threat to human health in light of the low livestock density and continued spread of epidemic hospital and community strains of MRSA in Canada remains unknown.
Dr Golding is a research scientist at the National Microbiology Laboratory, Winnipeg. His primary research interest focuses on antimicrobial drug resistance mechanisms, genomics, typing, and surveillance of S. aureus.
Acknowledgments
We thank Jennifer Campbell and Dave Spreitzer for performing PFGE; Brynn Kaplen, Claude Ouellette, and Erika Landry for performing amplicon cleanup and DNA sequencing; and Philip Mabon, Kunyan Zhang, and Lisa Louie for helping with analysis of the SCCmec element.
Funding was provided by the Federal Genomics Research and Development Initiative.
References
- Voss A, Loeffen F, Bakker J, Klaassen C, Wulf M. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg Infect Dis. 2005;11:1965–6.PubMedGoogle Scholar
- Khanna T, Friendship R, Dewey C, Weese JS. Methicillin resistant Staphylococcus aureus colonization in pigs and pig farmers. Vet Microbiol. 2008;128:298–303. DOIPubMedGoogle Scholar
- Smith TC, Male MJ, Harper AL, Kroeger JS, Tinkler GP, Moritz ED, Methicillin-resistant Staphylococcus aureus (MRSA) strain ST398 is present in midwestern U.S. swine and swine workers. PLoS One. 2009;4:e4258. DOIPubMedGoogle Scholar
- Golding GR, Campbell JL, Spreitzer DJ, Veyhl J, Surynicz K, Simor A, ; Canadian Nosocomial Infection Surveillance Program. A preliminary guideline for the assignment of methicillin-resistant Staphylococcus aureus to a Canadian pulsed-field gel electrophoresis epidemic type using spa typing. Can J Infect Dis Med Microbiol. 2008;19:273–81.PubMedGoogle Scholar
- Harmsen D, Claus H, Witte W, Rothgänger J, Claus H, Turnwald D, Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting using a novel software for spa-repeat determination and database management. J Clin Microbiol. 2003;41:5442–8. DOIPubMedGoogle Scholar
- Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–15.PubMedGoogle Scholar
- McDonald RR, Antonishyn NA, Hansen T, Snook LA, Nagle E, Mulvey MR, Development of a triplex real-time PCR assay for detection of Panton-Valentine leukocidin toxin genes in clinical isolates of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:6147–9. DOIPubMedGoogle Scholar
- Zhang K, McClure JA, Elsayed S, Louie T, Conly JM. Novel multiplex PCR assay for characterization and concomitant subtyping of staphylococcal cassette chromosome mec types I to V in methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2005;43:5026–33. DOIPubMedGoogle Scholar
- Kondo Y, Ito T, Ma XX, Watanabe S, Kreiswirth BN, Etienne J, Combination of multiplex PCRs for SCCmec type assignment: rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob Agents Chemother. 2007;51:264–74. DOIPubMedGoogle Scholar
- Mulvey MR, Chui L, Ismail J, Louie L, Murphy C, Chang N, ; Canadian Committee for the Standardization of Molecular Methods. Development of a Canadian standardized protocol for subtyping methicillin-resistant Staphylococcus aureus using pulsed-field gel electrophoresis. J Clin Microbiol. 2001;39:3481–5. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; 17th informational supplement. Document M100–S17. Wayne (PA): The Institute; 2007.
- Skov R, Frimodt-Moller N, Espersen F. Correlation of MIC methods and tentative interpretive criteria for disk diffusion susceptibility testing using NCCLS methodology for fusidic acid. Diagn Microbiol Infect Dis. 2001;40:111–6. DOIPubMedGoogle Scholar
- Walker ES, Levy F, Shorman M, David G, Abdalla J, Sarubbi FA. A decline in mupirocin resistance in methicillin-resistant Staphylococcus aureus accompanied administrative control of prescriptions. J Clin Microbiol. 2004;42:2792–5.
- Staden R, Beal KF, Bonfield JK. The Staden package. In: Misener S, Krawetz SA, editors. Computer methods in molecular biology, bioinformatics methods and protocols. Vol. 132. Totowa (NJ): The Humana Press Inc.; 1998. p. 115–130.
- Meyer F, Goesmann A, McHardy AC, Bartels D, Bekel T, Clausen J, GenDB—an open source genome annotation system for prokaryote genomes. Nucleic Acids Res. 2003;31:2187–95. DOIPubMedGoogle Scholar
- Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007 Jul;35(Web Server issue):W52–7.
- Schwarz S, Kalec K, Strommenger B. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius detected in the Bft-GermVet monitoring programme 2004–2006 in Germany. J Antimicrob Chemother. 2008;61:282–5. DOIPubMedGoogle Scholar
- van Duijkeren E, Jansen MD, Flemming SC, de Neeling H, Wagenaar JA, Schoormans AH, Methicillin-resistant Staphylococcus aureus in pigs with exudative epidermitis. Emerg Infect Dis. 2007;13:1408–10.PubMedGoogle Scholar
- Witte W, Strommenger B, Stanek C, Cuny C. Methicillin-resistant Staphylococcus aureus ST398 in humans and animals, Central Europe. Emerg Infect Dis. 2007;13:255–8. DOIPubMedGoogle Scholar
- Krziwanek K, Metz-Gercek S, Mittermayer H. Methicillin-resistant Staphylococcus aureus ST398 from human patients, upper Austria. Emerg Infect Dis. 2009;15:766–9. DOIPubMedGoogle Scholar
- Wulf MW, Sørum M, van Nes A, Skov R, Melchers WJ, Klaassen CH, Prevalence of methicillin-resistant Staphylococcus aureus among veterinarians: an international study. Clin Microbiol Infect. 2008;14:29–34. DOIPubMedGoogle Scholar
- van Rijen MM, Van Keulen PH, Kluytmans JA. Increase in a Dutch hospital of methicillin-resistant Staphylococcus aureus related to animal farming. Clin Infect Dis. 2008;46:261–3. DOIPubMedGoogle Scholar
- Gibbs SG, Green CF, Tarwater PM, Mota LC, Mena KD, Scarpino PV. Isolation of antibiotic-resistant bacteria from the air plume downwind of a swine confined or concentrated animal feeding operation. Environ Health Perspect. 2006;114:1032–7. DOIPubMedGoogle Scholar
- van Loo I, Huijsdens X, Tiemersma E, de Neeling A, van de Sande-Bruinsma N, Beaujean D, Emergence of methicillin-resistant Staphylococcus aureus of animal origin in humans. Emerg Infect Dis. 2007;13:1834–9.PubMedGoogle Scholar
- Simor AE, Ofner-Agostini M, Bryce E, McGeer A, Paton S, Mulvey MR; Canadian Hospital Epidemiology Committee and Canadian Nosocomial Infection Surveillance Program. Laboratory characterization of methicillin-resistant Staphylococcus aureus in Canadian hospitals: results of 5 years of national surveillance, 1995–1999. J Infect Dis. 2002;186:652–60. DOIPubMedGoogle Scholar
- Christianson S, Golding GR, Campbell J. Canadian Nosocomial Infection Surveillance Program, Mulvey MR. Comparative genomics of Canadian epidemic lineages of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2007;45:1904–11. DOIPubMedGoogle Scholar
- de Neeling AJ, van den Broek MJ, Spalburg EC, van Santen-Verheuvel MG, Dam-Deisz WDC, Boshuizen HC, High prevalence of methicillin resistant Staphylococcus aureus in pigs. Vet Microbiol. 2007;122:366–72. DOIPubMedGoogle Scholar
- Kunin V, Sorek R, Hugenholtz P. Evolutionary conservation of sequence and secondary structures in CRISPR repeats. Genome Biol. 2007;8:R61 . DOIPubMedGoogle Scholar
- Grissa I, Vergnaud G, Pourcel C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics. 2007;8:172. DOIPubMedGoogle Scholar
- Brouns SJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJ, Snijders AP, Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. DOIPubMedGoogle Scholar
- Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. DOIPubMedGoogle Scholar
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in staphylococci by targeting DNA. Science. 2008;322:1843–5. DOIPubMedGoogle Scholar
- Monecke S, Jatzwauk L, Weber S, Slickers P, Ehricht R. DNA microarray-based genotyping of methicillin-resistant Staphylococcus aureus strains from eastern Saxony. Clin Microbiol Infect. 2008;14:534–45. DOIPubMedGoogle Scholar
- Walther B, Monecke S, Ruscher C, Friedrich AW, Ehricht R, Slickers P, Comparative molecular analysis substantiates zoonotic potential of equine methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2009;47:704–10. DOIPubMedGoogle Scholar
- Godde JS, Bickerton A. The repetitive DNA elements called CRISPRs and their associated genes: evidence of horizontal transfer among prokaryotes. J Mol Evol. 2006;62:718–29. DOIPubMedGoogle Scholar
- Bhat M, Dumortier C, Taylor BS, Miller M, Vasquez G, Yunen J, Staphylococcus aureus ST398, New York City and Dominican Republic. Emerg Infect Dis. 2009;15:285–7. DOIPubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 16, Number 4—April 2010
| EID Search Options |
|---|
|
|
|
|
|
|

Please use the form below to submit correspondence to the authors or contact them at the following address:
Michael R. Mulvey, National Microbiology Laboratory, 1015 Arlington Ave, Winnipeg, Manitoba R3E 3R2, Canada
Top