Hypervirulent emm59 Clone in Invasive Group A Streptococcus Outbreak, Southwestern United States
David M. Engelthaler
1
, Michael Valentine
1, Jolene Bowers, Jennifer Pistole, Elizabeth M. Driebe, Joel Terriquez, Linus Nienstadt, Mark Carroll, Mare Schumacher, Mary Ellen Ormsby, Shane Brady, Eugene Livar, Del Yazzie, Victor Waddell, Marie Peoples, Kenneth Komatsu, and Paul Keim
Author affiliations: Translational Genomics Research Institute, Flagstaff, Arizona, USA (D. Engelthaler, M. Valentine, J. Bowers, E.M. Driebe, P. Keim); Arizona Department of Health Services, Phoenix, Arizona, USA (J. Pistole, S. Brady, E. Livar, V. Waddell, K. Komatsu); Northern Arizona Healthcare, Flagstaff (J. Terriquez, L. Nienstadt, M. Carroll); Coconino County Public Health Services District, Flagstaff (M. Schumacher, M.E. Ormsby, M. Peoples); Navajo Division of Health, Window Rock, Arizona, USA (D. Yazzie); Northern Arizona University, Flagstaff (P. Keim)
Main Article
Figure 1
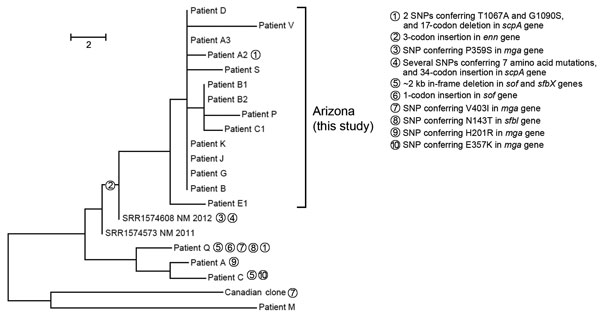
Figure 1. Phylogenetic single-nucleotide polymorphism (SNP) tree of emm59 isolates from a northern Arizona hospital displaying distribution of mutations in a 23kb positively selected region during invasive group A Streptococcus outbreak, southwestern United States. Maximum parsimony tree of all SNP loci (n = 58) in emm59 isolates (n = 18) from Arizona, 2 recent New Mexico isolate genomes, and the Canadian clone reference isolate MGAS15252. Consistency index = 1.0. Branch lengths represent numbers of SNPs between isolates; unit bar is in the figure. Numbered circles distinguish lineages of selected mutations in scpA, enn, sfbl, mga, sfbx, and sof genes in a 23-kb hotspot mutational region. Scale bar indicates SNPs.
Main Article
Page created: March 16, 2016
Page updated: March 16, 2016
Page reviewed: March 16, 2016
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
