Volume 22, Number 9—September 2016
Research
Staphylococcus aureus Regulatory RNAs as Potential Biomarkers for Bloodstream Infections
Figure 1
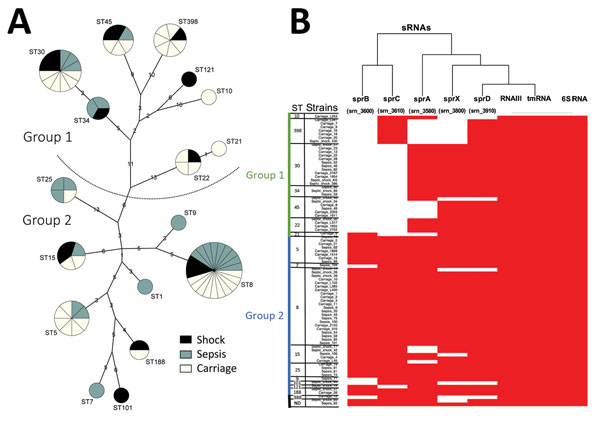
Figure 1. Phylogenetic and molecular typing of Staphylococcus aureus by using small RNAs (SRNAs), Rennes, France. A) Phylogenetic tree showing sRNA gene content and expression. Genotyping by multilocus sequence typing (MLST) and the maximum-parsimony method classified 83 S. aureus isolates (40 from patients with bloodstream infections and 40 from nasal carriers). Strains were distributed into 17 sequence types (STs) in groups 1 and 2 according to the method of Cooper and Feil (16). Three isolates were not included in the tree because 2 could not be typed and 1 belonged to the divergent ST883 lineage. Numbers indicate genetic distances between nodes. Circle sizes are proportional to number of isolates, and colors indicate origin. B) sRNA gene distribution among 83 analyzed strains. Five sRNAs were from pathogenicity islands and 3 sRNAs were from the core genome. Red indicates presence of sRNA and white indicates absence of SRNA. Heat map was constructed by using R software (https://www.r-project.org/). tmRNA, transfer–messenger RNA; ND, not determined.
References
- van Belkum A, Verkaik NJ, de Vogel CP, Boelens HA, Verveer J, Nouwen JL, Reclassification of Staphylococcus aureus nasal carriage types. J Infect Dis. 2009;199:1820–6 .DOIPubMedGoogle Scholar
- von Eiff C, Becker K, Machka K, Stammer H, Peters G. Nasal carriage as a source of Staphylococcus aureus bacteremia. Study Group. N Engl J Med. 2001;344:11–6 .DOIPubMedGoogle Scholar
- Smyth DS, Kafer JM, Wasserman GA, Velickovic L, Mathema B, Holzman RS, Nasal carriage as a source of agr-defective Staphylococcus aureus bacteremia. J Infect Dis. 2012;206:1168–77 .DOIPubMedGoogle Scholar
- Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev. 2015;28:603–61 .DOIPubMedGoogle Scholar
- Rosenstein R, Gotz F. What distinguishes highly pathogenic staphylococci from medium- and non-pathogenic? Curr Top Microbiol Immunol. 2013;358:33–89 .DOIPubMedGoogle Scholar
- Chabelskaya S, Gaillot O, Felden B. A Staphylococcus aureus small RNA is required for bacterial virulence and regulates the expression of an immune-evasion molecule. PLoS Pathog. 2010;6:e1000927 .DOIPubMedGoogle Scholar
- Romilly C, Lays C, Tomasini A, Caldelari I, Benito Y, Hammann P, A non-coding RNA promotes bacterial persistence and decreases virulence by regulating a regulator in Staphylococcus aureus. PLoS Pathog. 2014;10:e1003979 .DOIPubMedGoogle Scholar
- Sassi M, Augagneur Y, Mauro T, Ivain L, Chabelskaya S, Hallier M, SRD: a Staphylococcus regulatory RNA database. RNA. 2015;21:1005–17 .DOIPubMedGoogle Scholar
- Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541–64 .DOIPubMedGoogle Scholar
- Powers ME, Bubeck Wardenburg J. Igniting the fire: Staphylococcus aureus virulence factors in the pathogenesis of sepsis. PLoS Pathog. 2014;10:e1003871 .DOIPubMedGoogle Scholar
- Dellinger RP, Levy MM, Rhodes A, Annane D, Gerlach H, Opal SM, Surviving sepsis campaign: international guidelines for management of severe sepsis and septic shock, 2012. Intensive Care Med. 2013;39:165–228 .DOIPubMedGoogle Scholar
- Christensen S, Johansen MB, Christiansen CF, Jensen R, Lemeshow S. Comparison of Charlson Comorbidity Index with SAPS and APACHE scores for prediction of mortality following intensive care. Clin Epidemiol. 2011;3:203–11 .DOIPubMedGoogle Scholar
- Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38:1008–15.PubMedGoogle Scholar
- Chabelskaya S, Bordeau V, Felden B. Dual RNA regulatory control of a Staphylococcus aureus virulence factor. Nucleic Acids Res. 2014;42:4847–58 .DOIPubMedGoogle Scholar
- Dauwalder O, Lina G, Durand G, Bes M, Meugnier H, Jarlier V, Epidemiology of invasive methicillin-resistant Staphylococcus aureus clones collected in France in 2006 and 2007. J Clin Microbiol. 2008;46:3454–8 .DOIPubMedGoogle Scholar
- Cooper JE, Feil EJ. The phylogeny of Staphylococcus aureus: which genes make the best intra-species markers? Microbiology. 2006;152:1297–305 .DOIPubMedGoogle Scholar
- Robert J, Tristan A, Cavalie L, Decousser JW, Bes M, Etienne J, Panton-Valentine leukocidin–positive and toxic shock syndrome toxin 1–positive methicillin-resistant Staphylococcus aureus: a French multicenter prospective study in 2008. Antimicrob Agents Chemother. 2011;55:1734–9 .DOIPubMedGoogle Scholar
- Le Moing V, Alla F, Doco-Lecompte T, Delahaye F, Piroth L, Chirouze C, Staphylococcus aureus bloodstream infection and endocarditis: a prospective cohort study. PLoS One. 2015;10:e0127385 .DOIPubMedGoogle Scholar
- Feil EJ, Cooper JE, Grundmann H, Robinson DA, Enright MC, Berendt T, How clonal is Staphylococcus aureus? J Bacteriol. 2003;185:3307–16 .DOIPubMedGoogle Scholar
- Kiehn TE, Wong B, Edwards FF, Armstrong D. Comparative recovery of bacteria and yeasts from lysis-centrifugation and a conventional blood culture system. J Clin Microbiol. 1983;18:300–4.PubMedGoogle Scholar
- Keiler KC. Mechanisms of ribosome rescue in bacteria. Nat Rev Microbiol. 2015;13:285–97 .DOIPubMedGoogle Scholar
- Smith EJ, Visai L, Kerrigan SW, Speziale P, Foster TJ. The Sbi protein is a multifunctional immune evasion factor of Staphylococcus aureus. Infect Immun. 2011;79:3801–9 .DOIPubMedGoogle Scholar
- Hotchkiss RS, Monneret G, Payen D. Immunosuppression in sepsis: a novel understanding of the disorder and a new therapeutic approach. Lancet Infect Dis. 2013;13:260–8 .DOIPubMedGoogle Scholar
- Painter KL, Krishna A, Wigneshweraraj S, Edwards AM. What role does the quorum-sensing accessory gene regulator system play during Staphylococcus aureus bacteremia? Trends Microbiol. 2014;22:676–85 .DOIPubMedGoogle Scholar
- Burman JD, Leung E, Atkins KL, O’Seaghdha MN, Lango L, Bernado P, Interaction of human complement with Sbi, a staphylococcal immunoglobulin-binding protein: indications of a novel mechanism of complement evasion by Staphylococcus aureus. J Biol Chem. 2008;283:17579–93 .DOIPubMedGoogle Scholar
- Gonzalez CD, Ledo C, Giai C, Garofalo A, Gomez MI. The Sbi protein contributes to Staphylococcus aureus inflammatory response during systemic infection. PLoS One. 2015;10:e0131879 .DOIPubMedGoogle Scholar
- Foster TJ, Geoghegan JA, Ganesh VK, Hook M. Adhesion, invasion and evasion: the many functions of the surface proteins of Staphylococcus aureus. Nat Rev Microbiol. 2014;12:49–62 .DOIPubMedGoogle Scholar
- Rogasch K, Ruhmling V, Pane-Farre J, Hoper D, Weinberg C, Fuchs S, Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J Bacteriol. 2006;188:7742–58 .DOIPubMedGoogle Scholar
- Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005;5:751–62 .DOIPubMedGoogle Scholar
- Young BC, Golubchik T, Batty EM, Fung R, Larner-Svensson H, Votintseva AA, Evolutionary dynamics of Staphylococcus aureus during progression from carriage to disease. Proc Natl Acad Sci U S A. 2012;109:4550–5 .DOIPubMedGoogle Scholar
- Song J, Lays C, Vandenesch F, Benito Y, Bes M, Chu Y, The expression of small regulatory RNAs in clinical samples reflects the different life styles of Staphylococcus aureus in colonization vs. infection. PLoS One. 2012;7:e37294 .DOIPubMedGoogle Scholar