Volume 7, Number 3—June 2001
Research
Molecular Epidemiology of Serogroup A Meningitis in Moscow, 1969 to 1997
Abstract
Molecular analysis of 103 serogroup A Neisseria meningitidis strains isolated in Moscow from 1969 to 1997 showed that four independent clonal groupings were responsible for successive waves of meningococcal disease. An epidemic from 1969 to the mid-1970s was caused by genocloud 2 of subgroup III, possibly imported from China. Subsequent endemic disease through the early 1990s was caused by subgroup X and then by subgroup VI, which has also caused endemic disease elsewhere in Eastern Europe. A 1996 epidemic was part of the pandemic spread from Asia of genocloud 8 of subgroup III. Recent genocloud 8 epidemic disease in Moscow may represent an early warning for spread of these bacteria to other countries in Europe.
Until the mid-1970s, Neisseria meningitidis expressing the serogroup A capsular polysaccharide caused numerous epidemics of meningitis and septicemia, with annual incidence rates >100/100,000 (1). During serogroup A epidemics, these bacteria can also be isolated from the nasopharynx of a considerable proportion of the healthy population (2). After the mid-1970s, large serogroup A epidemics became rare, except in the so-called Meningitis Belt in the Sahel region of Africa (3,4). In 1997, N. meningitidis caused approximately 500,000 cases of disease and 50,000 deaths worldwide, half of them in Africa (5). Serogroup A meningococci are currently isolated only rarely in Western Europe and the United States, and a rise in either the healthy carriage rate or the proportion of serogroup A cases would be cause for concern.
Epidemic isolates of N. meningitidis have been assigned to a limited number of clonal groupings, historically by multilocus enzyme electrophoresis (MLEE) (4) and currently by multilocus sequence typing (MLST) (6). For MLST, sequences of housekeeping gene fragments are assigned different numeric allele designations, even if they differ by only one nucleotide. The combination of alleles from seven housekeeping gene fragments is called the sequence type (ST). Serogroup A bacteria have also been assigned to clonal groupings by random amplification of polymorphic DNA (RAPD) (7). These three methods yield largely concordant results, except that MLST is the most conservative and RAPD differentiates the largest number of isolates. The clonal groupings among serogroup A meningococci have been designated subgroups I through IX according to MLEE (7,8). Some of these subgroups have not been differentiated by RAPD analysis (I vs. II, III vs. VIII) (7) or MLST.
Recent epidemic serogroup A disease in Europe has largely reflected the pandemic patterns of spread of subgroup III meningococci. After a large subgroup III epidemic in China in the mid-1960s (8), these bacteria caused an outbreak of meningococcal disease in 1969 in western Norway (9), followed in the mid-1970s by a major epidemic in Finland (10). During the 1970s, subgroup III meningococci were isolated throughout western and northern Europe (11). In 1987, serogroup A, subgroup III meningococci caused a meningitis epidemic during the Hajj pilgrimage to Mecca, Saudi Arabia (12), and healthy pilgrims carried these bacteria throughout the world. Localized serogroup A disease was associated with former pilgrims for several years in the United Kingdom (13) and France (14), but these meningococci have since largely disappeared from western Europe. Rare endemic serogroup A disease was associated with subgroup VI in East Germany in the 1980s (8) and with subgroup IX in the Netherlands in the early 1990s (7).
Meningococci are naturally transformable and can import novel alleles from neisseriae that colonize the nasopharynx (15,16). However, these recombinant strains are usually lost during the bottlenecks caused by limited numbers of bacteria being transmitted during epidemic spread (17). Occasionally, variants pass through these bottlenecks first, and all subsequent progeny have a common genetic background different from that of the parental bacteria (17). Most subgroup III bacteria isolated from the mid-1960s to the mid-1980s ("pre-Mecca") had particular alleles of the hypervariable opaB and opaD loci encoding Opa (opacity) proteins and the iga locus encoding IgA1 protease. In contrast, subgroup III bacteria isolated after the 1987 Mecca epidemic ("post-Mecca") had other alleles at these three loci (17). Exceptional isolates with still other alleles at one or more of these loci were also found in several countries, but these variants all disappeared during subsequent epidemic spread (17).
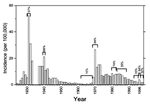
Data in the English literature about meningococci from Russia and other areas of eastern Europe are scarce. In recent decades, most patients with meningitis in Moscow (population 8.6 million during 1990 to 1999) have been treated at the Second Moscow Hospital for Infectious Diseases. Our review of their records shows that from 1980 to 1999, the annual incidence rates of meningitis caused by Haemophilus influenzae and Streptococcus pneumoniae were approximately 0.2/100,000 and 0.4/100,000, respectively. In contrast, several epidemics of meningococcal disease with much higher incidence rates occurred in Moscow from 1924 to 1998 (Figure 1). Large serogroup A epidemics occurred in 1931 and 1940, followed by a decline in disease incidence until the late 1960s. In 1968, a serogroup A epidemic began in Lipezk, Russia, where factory workers had recently arrived from Vietnam. These immigrants had traveled by rail through China, where subgroup III disease was prevalent (8). The incidence rate of meningococcal disease in Moscow and the proportion of serogroup A isolates remained high until the 1980s. In 1996, a new outbreak of serogroup A disease began, again initially associated with the Vietnamese community. During the outbreak peak in early 1996, 50 (34%) of 147 patients with meningococcal disease were Vietnamese, and all their disease isolates were serogroup A. During the rest of 1996, only 8 (6%) of 145 patients with meningococcal disease belonged to the Vietnamese community. During 1997 to 1999, hundreds of thousands of Muscovites were immunized with A polysaccharide vaccine. The incidence rate and the proportion of serogroup A isolates decreased and have remained stable through 1999.
We present data on the molecular epidemiology and genetic relationships of serogroup A meningococci isolated in Moscow from 1969 to 1997. The data show that four clonal groupings have been responsible for successive waves of disease and that epidemic disease in Moscow since 1994 is associated with a new subgroup III pandemic.
Serogroup A Isolates from Moscow
One hundred three strains of N. meningitidis were isolated in Moscow from the cerebrospinal fluid or blood of patients with systemic meningococcal disease or from throat swabs of their close contacts (seven subgroup VI strains, 1988-89). Strains were serogrouped by immune precipitation with specific sera and lyophilized. The bacteria we describe represent almost all the viable serogroup A bacteria still available in the laboratory collections at the Central Research Institute of Epidemiology and the Gamaleya Institute of Epidemiology and Microbiology, with the following exceptions: Only one third of the bacteria available from 1969 to 1972 and half the bacteria from 1996 were tested to avoid bias toward these two periods. The lyophilized bacteria were purified again by single colony isolation. These bacterial cultures were then serogrouped, serotyped, and subtyped by using whole-cell enzyme-linked immunosorbent assay (ELISA) with monoclonal antibodies (8). DNA was prepared as described (21) and used for molecular fine typing and MLST.
MLST
Both strands of fragments of abcZ, adk, aroE, gdh, pdhC, pgm, and fumC were sequenced as described (http://www.mlst.net), except that primers O1101 (5'TCCGGCTTGCCGTTTGTCAG) and O1102 (5'TTGTAGGCGGTTTTGGCGAC) were used for fumC. At the beginning of this project, the MLST database inadequately represented the diversity of serogroup A meningococci because it contained data for only 36 strains, predominantly from MLEE subgroups I, III, and IV-1 (6). To enlarge the database, MLST was performed with 66 serogroup A isolates from diverse sources that had previously been assigned to subgroups II through IX by MLEE (7,8) and in some cases by RAPD analysis (7). MLST was also performed with 42 serogroup A isolates from Moscow chosen from the most diverse branches of the RAPD tree. Other laboratories had deposited data for eight serogroup A strains in six novel STs, resulting in 152 serogroup A strains in 31 STs (Table 1). The results, including strain description, source, and MLEE assignments, are publicly available (http://www.mlst.net).
Molecular Fine Typing of Subgroup III
Alleles of the four chromosomal opa genes were determined by restriction analysis of polymerase chain reaction (PCR) products generated by using one primer in the upstream conserved flanking DNA and a second HV2-specific primer, as described (17). The complete opa genes were also sequenced from selected representative strains.
A fragment of iga was amplified from chromosomal DNA by using primers O103 and O115 and tested for a BfaI site that is present in iga1 but not in iga2 or iga3 (17). A different fragment of iga was amplified with primers O113 and O151 and tested for a DdeI site that is present in iga3 but not iga2 or iga1 (17). In addition, the entire iga gene was amplified with primers O103 and O152 and sequenced from one ST5 and one ST7 strain.
The IS1106A locus near opcA was amplified by using primers in conserved DNA flanking this locus (22). Two sizes of PCR products (4.8 and 3.5 kb) were obtained among the subgroup III strains. (One strain yielded a 12-kb product that was not investigated further.) An internal, polymorphic 307-bp stretch was sequenced from one product of each size and found to differ by 19 bp, including four restriction sites for HaeIII, ClaI, and BclI. The allele in the 4.8-kb fragment was named IS1106A10, and the allele in the 3.5-kb fragment was named IS1106A7. PCR products from all the subgroup III strains were tested for the four distinctive restriction sites, but no additional diversity was identified. Details on alleles at this locus in subgroup III bacteria from sources other than Moscow have been described (23).
The 2.1-kb tbpB gene was amplified by using an internal primer located near the ATG start codon (3'-Met2) and a primer located in the intergenic region between tbpB and tbpA (5'-Inter1), as described (16). Different alleles of tbpB were recognized by digesting the 2.1-kb fragment with HaeIII, MspI, and SspI and by sequencing the entire fragment from two ST5 strains and one ST7 strain. The tbpB allele designations are congruent with those described for a 600-bp 5'-terminal region that has been examined from numerous other strains (16).
To distinguish between pgm3 and pgm19 (6), the pgm locus was PCR-amplified by using primers O781 (5'-CGGCGATGCCGACCGCTTGG) and O782 (5'GGTGATGATTTCGGTTGCGCC). The PCR products were tested for an HinfI site that is absent in pgm3 and present in pgm19.
Genetic Relationships of Serogroup A Strains from Moscow
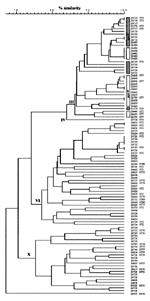
One hundred three serogroup A N. meningitidis strains isolated in Moscow during 1969 to 1998 were investigated by RAPD analysis to elucidate their genetic relationships. The data (Figure 2) revealed three primary clusters of isolates, two of which (subgroups III and VI) were assigned to known clonal groupings by comparison with previous data (7). The third cluster (24 isolates) differs from previously defined subgroups and was therefore tentatively designated subgroup X. Two singleton strains were labeled IV (a strain formerly assigned to subgroup IV-2 by MLEE) and ST79.
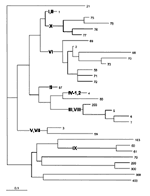
Forty-two strains that represent the diversity revealed by RAPD were analyzed by MLST. These data were combined with data from other strains as described in Materials and Methods, resulting in a database containing 152 serogroup A isolates (Table 1, Figure 3). MLST did not distinguish several subgroups defined by MLEE (I and II, IV-1 and IV-2, III and VIII, V and VII). Otherwise, the subgroup structure in a neighbor-joining (NJ) tree of the fraction of alleles differing between STs (Figure 3) was largely concordant with prior subgroup assignments from MLEE (8) or RAPD (7). Only STs 1 and 57 clustered inappropriately in the NJ tree (Figure 3).
The MLST results confirmed the assignments of isolates from Moscow to subgroups III and VI. They also demonstrated that subgroup X is distinct from previously defined subgroups and is closely related to subgroups I and II. Individual STs of these three subgroups share identical alleles for up to six of the seven gene fragments sequenced (Table 1).
Inconsistent Fine Typing by RAPD and MLST
Subgroup III isolates from Moscow were isolated during more than one epidemic wave, raising the question whether these isolates shared a common source. However, the concordance between RAPD and MLST did not extend below the subgroup level. The two major RAPD branches in subgroup III initially seemed to correspond to MLST ST5 and ST7. These STs are identical except for their pgm alleles (ST5, pgm3; ST7, pgm19,) which differ by 19 (4%) of 450 bp, including a distinctive restriction site. Genotyping of pgm from all subgroup III isolates by PCR-restriction fragment length polymorphism (PCR-RFLP) showed that both major RAPD branches contained both pgm alleles (Figure 2). Therefore, additional powerful fine typing methods, such as multilocus PCR-RFLP, were needed to resolve whether all bacteria within a subgroup originated from the same sources.
Serologic Properties
In some cases, serologic serotyping and serosubtyping can be used to enable rapid recognition of clonal groupings. The serologic properties of all the serogroup A bacteria from Moscow were determined by ELISA with monoclonal typing antibodies. The results confirmed previous results for subgroups III and VI (8) and showed that subgroup X is 4,21:P1.10 or 21:P1.10 (Table 2). Subgroup X is therefore not distinguishable by serologic testing from serogroup A subgroups I, II, IV-2 and VI, which are also 4,21:P1.10. The subgroup III strains uniformly expressed the class I and IIa pilin epitopes (8); however, the pilin epitopes of subgroups VI and X were not uniform: half of each group of strains expressed class I and IIa epitopes and the other half the class IIb epitope. Thus, except for subgroup III, serologic typing would not be useful for distinguishing these various subgroups.
Successive Waves Caused by Different Clonal Groupings
The three subgroups were isolated in Moscow during four successive waves of disease caused, respectively, by ST5 of subgroup III (1969 to 1977, 17 of 19 isolates), subgroup X (1983 to 1985; 21 of 27), subgroup VI (1986 to 1995; 32 of 36) and ST7 of subgroup III (1996-97; 19 of 21) (Table 3). The isolation of subgroup III bacteria correlated with epidemic peaks of disease in 1970 and 1996 (Figure 1); in both cases, subgroup III bacteria were also isolated in Moscow 1 to 2 years before the peak epidemic year (Table 3). In contrast, the isolation of subgroup X or subgroup VI did not correlate with an increase in the incidence rate for serogroup A disease. The slight increase in disease incidence in the mid-1980s was associated with serogroup B meningococci.
Molecular Fine Typing of Subgroup III Isolates
Both pre-Mecca and post-Mecca isolates are ST5, while the subgroup III meningococci isolated in Moscow since the mid-1990s are ST7. PCR-RFLP of the opaB, opaD and iga loci distinguished pre-Mecca from post-Mecca bacteria (17) but did not allow unambiguous assignment of the isolates from Moscow to either of these two waves. To resolve the fine structure of subgroup III, we identified three additional distinctive molecular markers, tbpB, IS1106A, and pgm, which were tested by PCR-RFLP plus sequencing for numerous subgroup III isolates from other countries (23). PCR-RFLP of these six variable loci subdivides subgroup III bacteria into nine so-called genoclouds, each containing a spectrum of related genotypes, and shows that a third wave of pandemic disease caused by genocloud 8 began in eastern Asia in the early 1990s. Genocloud 8 bacteria differ from other subgroup III meningococci in having the tbpB55, pgm19, and IS1106A7 alleles (Table 4). All 21 subgroup III bacteria isolated in Moscow from 1994 to 1997 also have these alleles (Table 4) and thus represent an extension of the third pandemic wave.
The subgroup III ST5 meningococci isolated in Moscow from 1969 to 1977 are distinct from all other subgroup III meningococci and define genocloud 2 (Table 4). The most closely related genocloud is genocloud 1, isolated in China in the mid-1960s, which contains the same IS1106A10 element but differs at tbpB and opaB. The genocloud 3 bacteria that caused outbreaks in northern Europe concurrently with the epidemic in Moscow differed in IS1106A (IS1106A4) as well as tbpB and opaB. Other subgroup III genoclouds, including genocloud 5 (post-Mecca), differed at even more loci (Table 4).
Phylogenetic Ancestry of Genocloud 2
The unique tbpB allele in ST5 subgroup III bacteria from Moscow is not informative for phylogenetic descent: It was not found in any other genocloud and was probably imported by DNA transformation (16) shortly before or after these bacteria arrived in Moscow. However, the phylogenetic history of the unique opaB alleles in the subgroup III bacteria from Moscow can be reconstructed from their sequences. Genocloud 1 bacteria from China have the opa132, opa92, opa131, and opa101 alleles at the opaA, B, D, and J loci, respectively. The novel alleles at the opaB and opaD loci in genocloud 2 bacteria from Moscow probably represent gene conversion with the opaJ101 and opaA132 alleles (Figure 4). The opaB140 allele, which was present in all but one strain from Moscow (Table 4), differs from opaJ101 by only one nucleotide (Figure 4). The opaB113 allele (one strain) can be explained by gene conversion of opaB140 with DNA from the opaA132 allele. opaD131 is present in genocloud 1 and was also found in all but two genocloud 2 strains from Moscow. The two exceptional strains contain the opaD109 and opaD110 alleles, which could easily have arisen by gene conversion of part of the opaD131 locus with DNA from the opaJ101 locus.
Since the 1920s, serogroup A meningococci have been responsible for a large proportion of the bacterial meningitis in Moscow (Figure 1). Serogroup A meningococci isolated in Moscow from 1969 to 1997 (103 isolates) were analyzed by RAPD. Strains representing the diversity revealed by the RAPD data were analyzed by MLST (44 isolates), and all 38 subgroup III strains were subjected to molecular fine typing by multilocus PCR-RFLP. The results illustrate the limitations of these methods and yield important information about the phylogenetic relationships among serogroup A meningococci. They also demonstrate that serogroup A meningococcal disease in Moscow since 1969 has been caused by four successive, independent waves of bacterial colonization, most recently by the spread of a third subgroup III pandemic wave from East Asia.
Typing Methods
RAPD analysis was used to provide a quick overview of the genetic diversity in the 103 strains investigated (Figure 2). This information was used to select a smaller group of bacteria to be tested by more expensive and labor-intensive methods such as MLST. As in previous comparisons between RAPD and MLEE (7), the subgroup assignments were concordant between MLST and RAPD. The RAPD data allowed the initial recognition of a new subgroup, subgroup X, and the MLST data confirmed that subgroup IX is not genetically related to the epidemic serogroup A subgroups as originally demonstrated by RAPD data (7). Thus, RAPD analysis can be a highly reliable, quick method for recognizing genetically related bacterial groups.
Finer subdivisions in subgroups III and X, suggested by the RAPD data, were contradicted by MLST results. ST76 was distributed among both primary RAPD subdivisions of the subgroup X cluster, and the two primary RAPD subdivisions of the subgroup III cluster did not correlate with presence of pgm3 versus pgm19 (Figure 2) or with epidemiologic data. Such random noise will probably be most extreme for bacteria such as N. meningitidis, which frequently import DNA from unrelated neisseriae (15). Thus, putative subdivisions within phylogenetically related groups suggested by RAPD data should be confirmed by independent methods.
MLST is based on sequences of conserved housekeeping genes and sequence variants are rarer than with most other methods used in molecular epidemiology, thus facilitating global, long-term epidemiology (6). For subgroup III, the distinction between STs 5 and 7 based on pgm is a very useful molecular marker for recognizing bacteria from the third pandemic wave. However, the epidemiologic data do not suggest that other STs are useful markers for genetic variants that have spread extensively, and MLST does not necessarily provide the discrimination desired for fine typing.
Unfortunately, the fine structure of bacteria that frequently import foreign DNA is very difficult to resolve, and we know of no method that is eminently suitable for this task. Our data argue against the use of RAPD and MLST. Although pulsed-field gel electrophoresis is highly suitable for differentiating almost identical bacteria in limited numbers of strains (17,24,25), it is poorly suited for elucidating the relationships among large numbers of less strongly related bacteria. Fine typing was therefore attempted based on multilocus RFLP of PCR fragments of pgm plus the variable genes opa, iga, and tbpB and the IS1106A element. Multilocus PCR-RFLP of variable genes allowed molecular fine typing within subgroup III meningococci, but the data are also limited by considerable random noise, resulting in genoclouds of related variants (23). Alternative strategies involving IS elements (26), amplified fragment-length polymorphism (27), or variable nucleotide repeat sequences (28) have been used successfully for fine typing of highly uniform microorganisms. Such methods have not yet been extensively tested with N. meningitidis but seem likely to be limited by at least as much random noise as the methods we used. Thus, reliable fine typing remains difficult to achieve.
Phylogeny of Serogroup A Meningococci
The MLST data on 152 serogroup A strains allow inferences about the evolution of these bacteria. The STs in subgroups I through VIII and X (the epidemic subgroups) probably stem from a common ancestor. They form a monophyletic clade in phylogenetic trees (Figure 3), and many of these STs share the fumC1, abcZ1, adk3, gdh1, pdhC1, and pgm3 alleles (Table 1). The epidemic subgroups include meningococci isolated since the beginning of this century (ST4, subgroup IV-2, 1917; ST57, subgroup II, 1930) (8) and must have evolved before then.
Older isolates of subgroups III (1960s), IV-1 (1960s), and IV-2 (1917) had identical or nearly identical opa, iga, and tbpB alleles (16,17), whereas more recent isolates have imported novel alleles at these loci. These data indicate that the common ancestor of these three subgroups existed approximately 100 years ago (17).
The branch lengths (Figure 3) and common alleles indicate that the epidemic subgroups are probably not much older than the split between subgroups III, IV-1, and IV-2. The epidemic serogroup A subgroups may therefore have descended from a common ancestor that existed within the last few hundred years. Epidemic meningitis was first observed in Geneva in 1805 (29), and that epidemic may have marked the evolution of serogroup A after the import of the sacA-D genes encoding the A polysaccharide. Horizontal exchange between serogroups B and C of genes encoding capsular polysaccharides has been documented (30), and more than one capsular polysaccharide serogroup is present in several of the otherwise uniform STs in the complete MLST database. Furthermore, the sacA-D genes are characterized by an unusually low GC content (31), as if these genes were recently imported from an unrelated species. If this speculation is correct, the other serogroup A bacteria (Figure 3, bottom) may represent unrelated N. meningitidis that have imported sacA-D genes from the epidemic subgroups. Alternatively, the sacA-D genes may have been imported from an unrelated species more than once, or the epidemic subgroups may have imported the sacA-D from one of the nonepidemic STs (Figure 3, bottom). Analysis of the sequence diversity of the sacA-D genes in various STs might resolve these alternative explanations.
Molecular Epidemiology
Evaluation of the long-term molecular epidemiology of bacterial pathogens within a global context is complex, requiring large, representative strain collections and reliable, preferably phylogenetically based, typing schemes. Our results illustrate both strengths and weaknesses in our knowledge of serogroup A epidemiology.
The assignment of genocloud 8 bacteria isolated in the mid-1990s in Moscow to the third subgroup III pandemic wave is straightforward and indisputable. It rests on three unusual markers (pgm19, tbpB55, and IS1106A7), and no differences in these three markers were detected among recent subgroup III isolates from China, Mongolia, Africa, and Russia. The initial association of genocloud 8 disease in Moscow with the Vietnamese community remains unexplained.
The genocloud 2 subgroup III bacteria from the early 1970s differ at the opaB and tbpB loci from all known subgroup III meningococci. We argue that genocloud 2 is probably derived from genocloud 1 in China because of the shared IS1106A10 allele, which was not found in any other subgroup III bacteria. In support of this interpretation based on molecular data, this epidemic first broke out among Vietnamese immigrants who had traveled through China en route to Russia. However, few subgroup III strains exist that were isolated before 1969, and our knowledge about the diversity and patterns of serogroup A disease before 1969 is only fragmentary.
Subgroup VI bacteria of serogroups A, B, and C were occasionally isolated from endemic disease in East Germany in the 1980s (8). Our data show that most of the serogroup A meningococci isolated during 1986 to 1994 in Moscow were also subgroup VI, and these bacteria may have been widely distributed in Eastern Europe in the 1980s. In contrast, subgroup X, common in Moscow during 1983 to 1985, has not been described previously. Various STs (Figure 3, bottom) have also been isolated only from single countries (subgroup IX in the Netherlands during 1989 to 1993 [7]; STs 299, 300, 388, and 400 in Czechoslovakia in 1972 [K. Jolley and P. Kriz, pers. comm.]; ST79 in Moscow in 1995; and ST103 in Greece in 1996 [R. Urwin, pers. comm.]). These bacteria have apparently not spread extensively, unlike the epidemic subgroups. In this respect, they resemble most endemic serogroup B meningococci, such as those identified by MLEE among endemic disease isolates from the Netherlands (32) and carrier isolates from Norway (33). Additional serogroup A STs specific to certain countries or regions probably remain to be identified. Current meningococcal strain collections containing a few thousand isolates are probably one or two orders of magnitude too small to yield reliable epidemiologic data for unusual STs that have not caused larger disease outbreaks.
Subgroup III has caused large disease outbreaks, and its molecular epidemiology has been extensively investigated (11,17,23). As a result, the available information on genetic changes during pandemic spread of these bacteria is among the most extensive for any bacterial pathogen. Our data show that the extensive pandemic spread of these bacteria is likely to continue. Although Moscow and western Europe differ somewhat in epidemiologic patterns, the recent extension of this third pandemic wave to Moscow warns us that we should be prepared for new, large serogroup A outbreaks in countries in both Eastern and western Europe.
Dr. Achtmann is a scientific group leader at the Max-Planck Institut für Infektionsbiologie, Berlin. His group investigates the population structures of bacterial pathogens, including N. meningitidis, Helicobacter pylori, and Yersinia pestis. These analyses integrate results from epidemiologic studies of global disease patterns.
Acknowledgments
We gratefully acknowledge information on sources of strains in the MLST database and permission to cite these data from Keith Jolley and Rachel Urwin.
This work was partly supported by grants Ac 36/9-2 from the Deutsche Forschungsgemeinschaft and INTAS.
References
- Peltola H. Meningococcal disease: still with us. Rev Infect Dis. 1983;5:71–91. DOIPubMedGoogle Scholar
- Crowe BA, Wall RA, Kusecek B, Neumann B, Olyhoek T, Abdillahi H, Clonal and variable properties of Neisseria meningitidis isolated from cases and carriers during and after an epidemic in the Gambia, West Africa. J Infect Dis. 1989;159:686–700. DOIPubMedGoogle Scholar
- Achtman M. Molecular epidemiology of epidemic bacterial meningitis. Rev Med Microbiol. 1990;1:29–38.
- Achtman M. Global epidemiology of meningococcal disease. In: Cartwright K, editor. Meningococcal disease. Chichester, England: John Wiley & Sons, Ltd; 1995:159-75.
- Tikhomirov E, Santamaria M, Esteves K. Meningococcal disease: public health burden and control. World Health Stat Q. 1997;50:170–6.PubMedGoogle Scholar
- Maiden MCJ, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A. 1998;95:3140–5. DOIPubMedGoogle Scholar
- Bart A, Schuurman IGA, Achtman M, Caugant DA, Dankert J, van der Ende A. Random amplified polymorphic DNA (RAPD) genotyping of serogroup A meningococci yields similar results to multilocus enzyme electrophoresis (MEE) and reveals new genotypes. J Clin Microbiol. 1998;36:1746–9.PubMedGoogle Scholar
- Wang J-F, Caugant DA, Li X, Hu X, Poolman JT, Crowe BA, Clonal and antigenic analysis of serogroup A Neisseria meningitidis with particular reference to epidemiological features of epidemic meningitis in China. Infect Immun. 1992;60:5267–82.PubMedGoogle Scholar
- Hjetland R, Caugant DA, Hofstad T, Frøholm LO, Selander RK, Serogroup A. Neisseria meningitidis of clone III-1 in western Norway, 1969-73. Scand J Infect Dis. 1990;22:241–2. DOIPubMedGoogle Scholar
- Mäkelä PH, Peltola H, Käyhty H, Jousimies H, Pettay O, Ruoslahti E, Polysaccharide vaccines to group A Neisseria meningitidis and Haemophilus influenzae type b: a field trial in Finland. J Infect Dis. 1977;136:43–50. DOIPubMedGoogle Scholar
- Achtman M, Kusecek B, Morelli G, Eickmann K, Wang J, Crowe B, A comparison of the variable antigens expressed by clone IV-1 and subgroup III of Neisseria meningitidis serogroup A. J Infect Dis. 1992;165:53–68. DOIPubMedGoogle Scholar
- Moore PS, Reeves MW, Schwartz B, Gellin BG, Broome CV. Intercontinental spread of an epidemic group A Neisseria meningitidis strain. Lancet. 1989;ii:260–3. DOIPubMedGoogle Scholar
- Jones DM, Sutcliffe EM. Group A meningococcal disease in England associated with the Haj. J Infect. 1990;21:21–5. DOIPubMedGoogle Scholar
- Riou JY, Caugant DA, Selander RK, Poolman JT, Guibourdenche M, Collatz E. Characterization of Neisseria meningitidis serogroup A strains from an outbreak in France by serotype, serosubtype, multilocus enzyme genotype and outer membrane protein pattern. Eur J Clin Microbiol Infect Dis. 1991;10:405–9. DOIPubMedGoogle Scholar
- Maiden MCJ, Malorny B, Achtman M. A global gene pool in the neisseriae. Mol Microbiol. 1996;21:1297–8. DOIPubMedGoogle Scholar
- Linz B, Schenker M, Zhu P, Achtman M. Frequent interspecific genetic exchange between commensal neisseriae and Neisseria meningitidis. Mol Microbiol. 2000;36:1049–58. DOIPubMedGoogle Scholar
- Morelli G, Malorny B, Müller K, Seiler A, Wang J, del Valle J, Clonal descent and microevolution of Neisseria meningitidis during 30 years of epidemic spread. Mol Microbiol. 1997;25:1047–64. DOIPubMedGoogle Scholar
- Bolshakov KN. Epidemiological and geographical aspects of meningococcal infection [dissertation]. Moscow: Moscow Univ.; 1973.
- Kostyukova NN, Mironova TK, Leschinskaya EV. Microbiological characteristics of meningococcus infection in Moscow during a periodic rise in morbidity (1968-1971). Zh Mikrobiol Epidemiol Immunobiol. 1974;5:29–32.
- Demina AA, Martinov JV. An analysis of the epidemic cycle of meningococcal infection in Russia (1969-1993). Zh Mikrobiol Epidemiol Immunobiol. 1996;4:57–61.PubMedGoogle Scholar
- Sarkari J, Pandit N, Moxon ER, Achtman M. Variable expression of the Opc outer membrane protein in Neisseria meningitidis is caused by size variation of a promoter containing poly-cytidine. Mol Microbiol. 1994;13:207–17. DOIPubMedGoogle Scholar
- Zhu P, Morelli G, Achtman M. The opcA and opcB regions in Neisseria: genes, pseudogenes, deletions, insertion elements and DNA islands. Mol Microbiol. 1999;33:635–50. DOIPubMedGoogle Scholar
- Zhu P, van der Ende A, Falush D, Beieske N, Morelli G, Linz B, Fit genotypes and escape variants of subgroup III Neisseria meningitidis during three pandemics of epidemic meningitis. Proc Natl Acad Sci U S A. 2001. In press.PubMedGoogle Scholar
- Kwara A, Adegbola RA, Corrah PT, Weber M, Achtman M, Morelli G, Meningitis caused by a serogroup W135 clone of the ET-37 complex of Neisseria meningitidis in West Africa. Trop Med Int Health. 1998;3:742–6. DOIPubMedGoogle Scholar
- Vogel U, Morelli G, Zurth K, Claus H, Kriener E, Achtman M, Necessity of molecular techniques to distinguish between Neisseria meningitidis isolated from meningococcal disease and from their healthy contacts. J Clin Microbiol. 1998;36:2465–70.PubMedGoogle Scholar
- Van Embden JDA, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendation for a standardized methodology. J Clin Microbiol. 1993;31:406–9.PubMedGoogle Scholar
- Keim P, Kalif A, Schupp J, Hill K, Travis SE, Richmond K, Molecular evolution and diversity in Bacillus anthracis as detected by amplified fragment length polymorphism markers. J Bacteriol. 1997;179:818–24.PubMedGoogle Scholar
- Keim P, Klevytska AM, Price LB, Schupp JM, Zinser G, Smith KL, Molecular diversity in Bacillus anthracis. J Appl Microbiol. 1999;87:215–7. DOIPubMedGoogle Scholar
- Vieusseux M. Mémoire sur le maladie qui a régné à Genève au printemps de 1805. J Med Chirurg Pharm. 1806;11:163–82.
- Swartley JS, Marfin AA, Edupuganti S, Liu LJ, Cieslak P, Perkins B, Capsule switching of Neisseria meningitidis. Proc Natl Acad Sci U S A. 1997;94:271–6. DOIPubMedGoogle Scholar
- Parkhill J, Achtman M, James KD, Bentley SD, Churcher C, Klee SR, Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature. 2000;404:502–6. DOIPubMedGoogle Scholar
- Scholten RJPM, Bijlmer HA, Poolman JT, Kuipers B, Caugant DA, van Alphen L, Meningococcal disease in the Netherlands, 1958-1990: a steady increase in the incidence since 1982 partially caused by new serotypes and subtypes of Neisseria meningitidis. J Infect Dis. 1993;16:237–46. DOIPubMedGoogle Scholar
- Caugant DA, Høiby EA, Magnus P, Scheel O, Hoel T, Bjune G, Asymptomatic carriage of Neisseria meningitidis in a randomly sampled population. J Clin Microbiol. 1994;32:323–30.PubMedGoogle Scholar
Figures
Tables
Cite This ArticleTable of Contents – Volume 7, Number 3—June 2001
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Mark Achtman, Max-Planck Institut für Infektionsbiologie, Schumannstr. 21/22, 10117 Berlin, Germany; fax: +49 30 28460 750
Top