Volume 11, Number 7—July 2005
Research
Nipah Virus in Lyle's Flying Foxes, Cambodia
Figure 2
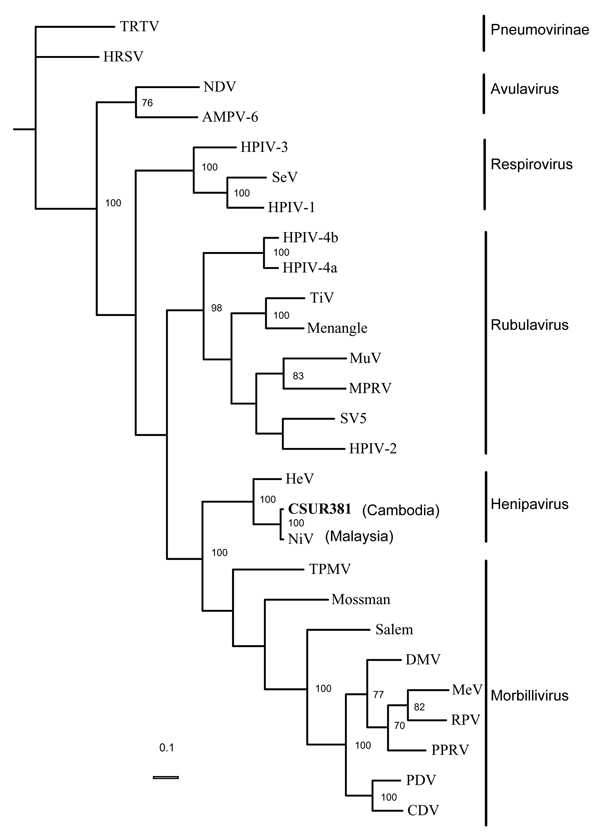
Figure 2. . Phylogenetic analysis of the 1,599 nucleotides of the N gene coding domain sequence from the Nipah virus Cambodian isolate, members of the subfamily Paramyxovirinae, and 2 species of the subfamily Pneumovirinae used as outgroups. GenBank accessions numbers used are as follows: APMV-6: Avian paramyxovirus 6, AY029299; CDV: Canine distemper virus, AF014953; DMV, Dolphin distemper virus, X75961; HeV: Hendra virus, AF017149; HPIV-1: Human parainfluenza virus 1, D011070; HPIV-2: Human parainfluenza virus 2, M55320; HPIV-3: Human parainfluenza virus 3, D10025; HPIV4a: Parainfluenza virus type 4A, M32982; HPIV4b: Parainfluenza virus type 4B, M32983; HRSV: Human respiratory syncytial virus, X00001; MeV: Measles virus, K01711; MPRV: Mapuera virus, X85128; Menangle: Menangle virus, AF326114; Mossman: Mossman virus, AY286409; MuV: Mumps virus, D86172; NDV: Newcastle disease virus, AF064091; NiV: Nipah virus, AF212302; PDV: Phocid distemper virus, X75717; PPRV, Peste des Petits ruminants virus, X74443; RPV: Rinder pest virus, X68311; Salem: Salem virus, AF237881; SeV: Sendai virus, X00087; SV5: Simian virus 5, M81442; TiV: Tioman virus, AF298895; TPMV: Tupaia paramyxovirus, AF079780; TRTV: Turkey Rhinotracheitis virus, AY640317. Significant bootstrap values (≥70%) are indicated. The phylogram was generated by parsimony method and analyzing 100 bootstrap replicates.