Volume 12, Number 11—November 2006
Letter
Chikungunya Fever, Hong Kong
Figure A2
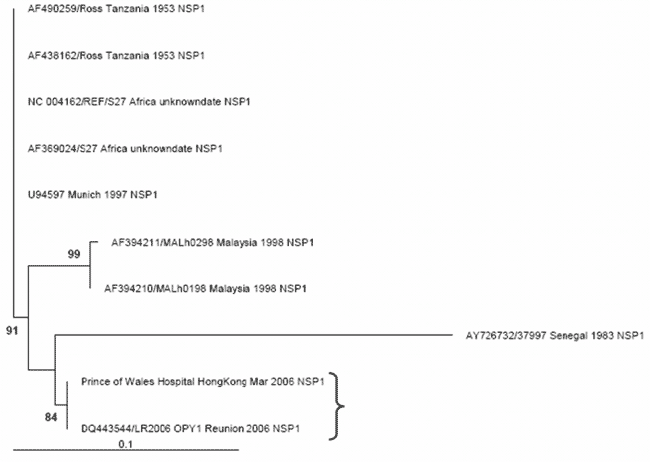
Figure A2. The maximum likelihood phylogenetic tree for chikungunya nonstructural protein 1 (NSP1) sequences (314 bp) constructed using PAUP* under a transitional model with equal base frequencies with a γ-distributed rate of substitution (i.e., TIM+G), as selected by MODELTEST (version 3.7) under the Akaike information criteria. The bracket shows that the PWH patient NSP1 sequence (DQ489788) clusters most closely with the outbreak La Reunion sequence (LR2006_OPY1_Reunion_2006_NSP1, DQ443544). Only bootstrap values >70 are shown and considered significant. The tree is rooted against the earliest sequence (AF490259/Ross_Tanzania_1953).
Page created: October 14, 2011
Page updated: October 14, 2011
Page reviewed: October 14, 2011
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.