Volume 12, Number 5—May 2006
Letter
Novel Recombinant Norovirus in China
Figure
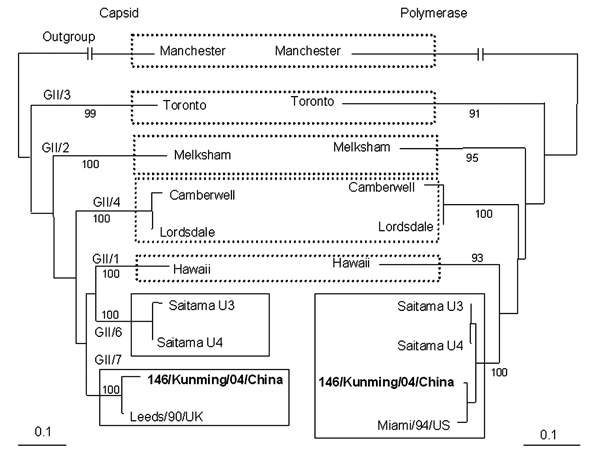
Figure. Changes in norovirus (NoV) genotypes on the basis of phylogentic trees of nucleotide sequences of 146/Kunming/04/China. Trees were constructed from partial nucleotide sequences of capsid and polymerase regions of 146/Kunming/04/China. 146/Kunming/04/China is boldface. Dashed boxes indicate the maintenance of genotypes of reference NoV strains, and solid boxes indicate the involvement of NoV genotypes with recombinant NoV 146/Kunming/04/China. A phylogenetic tree with 100 bootstrap resamples of the nucleotide alignment datasets was generated by using the neighbor-joining method with ClustalX. The genetic distance was calculated by using the Kimura 2-parameter method (PHYLIP). The scale indicates nucleotide substitutions per position. The numbers in the branches indicate the bootstrap values. Manchester strain was used as an outgroup strain for phylogenetic analysis. The nucleotide sequence of NoV strain 146/Kunming/04/China had been submitted to GenBank and has been assigned accession no. DQ304651. Reference NoV strains and accession nos. used in this study are as follows: Manchester (X86560), Toronto (U02030), Melksham (X81879), Camberwell (AF145896), Leeds/90/UK (AJ277608), Lordsdale (X86557), Hawaii (U07611), Saitama U3 (AB039776), Saitama U4 (AB039777), and Miami/94/US (AF414410).