Volume 16, Number 9—September 2010
Letter
Family Outbreak of Shiga Toxin–producing Escherichia coli O123:H–, France, 2009
To the Editor: Shiga toxin–producing Escherichia coli (STEC) is a major cause of foodborne disease in industrialized countries. We present results of the investigation of a family outbreak in France caused by a rare STEC serotype.
Surveillance of STEC infections in France since 1996 has been based on national surveillance of STEC-related pediatric hemolytic uremic syndrome (HUS) (1). On February 11, 2009, two cases of diarrhea were reported to a surveillance coordinator: 1 in a child with HUS and the other in that child’s sibling.
The 2 siblings, 2 and 6 years of age, had diarrhea beginning on February 4 and 5, 2009. Bloody diarrhea developed in the younger child, and HUS was diagnosed on February 9. The older child had nonbloody diarrhea for 3 days and abdominal pain. Questioning of the patients’ parents identified no recent history of travel, contact with farm animals, or outdoor bathing. A food history indicated that the 2 patients had shared an undercooked ground beef burger 4–5 days before symptom onset. The patients’ parents also ate burgers from the same package (box); they did not report any gastrointestinal symptoms.
Fecal specimens of the patients were tested for STEC by direct PCR for STEC genes (stx); after which culture and identification of stx1, stx2, eae, and ehxA (hlyA) virulence genes; and serotyping with a panel of 22 serum samples were conducted as described (1,2). Molecular serotyping was subsequently conducted on nonagglutinating strains by using the rfb–restriction fragment length polymorphism technique for O antigen (3) and sequencing of the fliC gene for H antigen (4).
A trace-back investigation was conducted for the implicated beef burgers, which were obtained from a box of 10, frozen, 100-g ground beef burgers purchased in late January 2009. The remaining beef burger in the box from which the patients had eaten a beef burger was obtained from the family’s freezer for microbiologic testing. Stored production samples from the implicated batch underwent microbiologic testing.
After broth enrichment, ground beef samples were tested by PCR for stx and eae virulence genes and O antigens of serotypes O157, O26, O145, O103, and O111 (2,5,6). Subsequently, strains isolated from stx-positive and eae-positive enrichment broths were biochemically tested and underwent serotyping and PCR identification of virulence genes. Genetic relatedness of clinical and ground beef STEC strains was studied by using pulsed-field gel electrophoresis with Xbal as described (7).
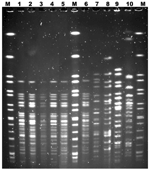
A nonmotile strain of STEC stx2 eae ehxA, which was not serotypeable by the panel of 22 serum samples, was identified in fecal samples from patients and in the remaining ground beef. Molecular serotyping of clinical isolates and an isolate from the beef identified a strain of STEC O123:H2. Analysis by pulsed-field gel electrophoresis indicated that the clinical and meat isolates were genetically related (Figure). The level of STEC contamination in the meat was 30–40 CFU/g. All stored meat production samples tested were negative for STEC.
A clinical strain and a ground beef STEC strain were sent to the World Health Organization Collaborating Centre for Reference and Research on Escherichia and Klebsiella in Copenhagen, Denmark, in December 2009 for analysis. The clinical strain was confirmed as STEC O123:H–, and the meat strain was confirmed as a nonmotile STEC rough type by serum agglutination. Both strains had virulence genes stx2a, eae, and ehxA (F. Scheutz, pers. comm.).
We identified a family outbreak of STEC O123:H– stx2a, eae ehxA infections associated with ingestion of undercooked ground beef. No similar cases of STEC infection were identified by active case finding. This serotype is rarely described as a cause of human clinical infection. No human isolate of serotype O123:H– is recorded in the database of the World Health Organization Collaborating Centre for Reference and Research on Escherichia and Klebsiella (F. Scheutz, pers. comm.).
Two strains of STEC O123:H– stx2d were isolated from asymptomatic persons in Germany during 1996–2000 (8). A study in Australia in 2003 reported using a strain of O123:H– stx1 stx2 ehxA from Switzerland that had been isolated from a person with diarrhea (9).
We report foodborne transmission of STEC O123:H– that resulted in a cluster of clinical cases of infection. Eating ground beef is a well-established mode of STEC transmission, particularly for serotype O157:H7. STEC serotype O123:H– has been isolated from feces of healthy lambs and sheep in Spain (10) and in southwestern Australia (9) and is considered to be among the predominant ovine STEC serotypes in these countries.
This family outbreak shows that STEC serotype O123:H–, albeit rarely described as causing human illness, can cause severe human infection. This serotype can also cause clusters of STEC infections and be transmitted by ingestion of undercooked ground beef.
Acknowledgment
We thank Sylvie Peters, Hélène Perin, Lydie Pachtchenko-Claudet, Marie-Odile Klippenspies, Julie Hanot, Laurent Claudet, Philippe Couratier, and Valerie le Bourg for assistance with investigating this outbreak; and Flemming Scheutz for verifying serotypes and virulence genes.
References
- Espié E, Grimont F, Mariani-Kurkdjian P, Bouvet P, Haeghebaert S, Filliol I, Surveillance of hemolytic uremic syndrome in children less than 15 years of age, a system to monitor O157 and non-O157 Shiga toxin–producing Escherichia coli infections in France, 1996–2006. Pediatr Infect Dis J. 2008;27:595–601. DOIPubMedGoogle Scholar
- Paton AW, Paton JC. Detection and characterization of Shiga toxigenic Escherichia coli by using multiplex PCR assays for stx1, stx2, eaeA, enterohemorrhagic E. coli hlyA, rfbO111, and rfbO157. J Clin Microbiol. 1998;36:598–602.PubMedGoogle Scholar
- Coimbra RS, Grimont F, Lenormand P, Burguière P, Beutin L, Grimont PAD. Identification of Escherichia coli O-serogroupes by restriction of the amplified O-antigen gene cluster (rfb-RFLP). Res Microbiol. 2000;151:639–54. DOIPubMedGoogle Scholar
- Coimbra RS, Lefevre M, Grimont F, Grimont PA. Clonal relationships among Shigella serotypes suggested by cryptic flagellin gene polymorphism. J Clin Microbiol. 2001;39:670–4. DOIPubMedGoogle Scholar
- Read SC, Clarke RC, Martin A, De Grandis SA, Hii J, McEwen S, Polymerase chain reaction for detection of verocytotoxigenic Escherichia coli isolated from animals and food sources. Mol Cell Probes. 1992;6:153–61. DOIPubMedGoogle Scholar
- Perelle S, Dilasser F, Grout J, Fach P. Detection by 5′-nuclease PCR of Shiga-toxin producing Escherichia coli O26, O55, O91, O103, O111, O113, O145 and O157:H7, associated with the world’s most frequent clinical cases. Mol Cell Probes. 2004;18:185–92. DOIPubMedGoogle Scholar
- Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67. DOIPubMedGoogle Scholar
- Friedrich AW, Bielaszewska M, Zhang WL, Pulz M, Kuczius T, Ammon A, Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. J Infect Dis. 2002;185:74–84. DOIPubMedGoogle Scholar
- Brett KN, Ramachandran V, Hornitzky MA, Bettelheim KA, Walker MJ, Djordjevic SP. stx1c is the most common Shiga toxin 1 subtype among Shiga toxin–producing Escherichia coli isolates from sheep but not among isolates from cattle. J Clin Microbiol. 2003;41:926–36. DOIPubMedGoogle Scholar
- Blanco M, Blanco JE, Mora A, Rey J, Alonso JM, Hermoso M, Serotypes, virulence genes, and intimin types of Shiga toxin (verotoxin)–producing Escherichia coli isolates from healthy sheep in Spain. J Clin Microbiol. 2003;41:1351–6. DOIPubMedGoogle Scholar
Figure
Cite This ArticleRelated Links
Table of Contents – Volume 16, Number 9—September 2010
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Lisa A. King, Département de Maladies Infectieuses, Institut de Veille Sanitaire, 12 rue du Val d’Osne, 94415 Saint-Maurice CEDEX, France
Top